CASA Guides:ATCA Advanced Continuum Polarization Tutorial NGC612-CASA4.7: Difference between revisions
No edit summary |
|||
| Line 212: | Line 212: | ||
[[File:Screenshot_plotms_ngc612_flag.png|thumb|Figure 1: Plotms displaying the worst RFI spikes]] | [[File:Screenshot_plotms_ngc612_flag.png|thumb|Figure 1: Plotms displaying the worst RFI spikes]] | ||
It is always a good idea to examine the data before jumping straight into calibration. Start by flagging data known to be bad from observing logs, then examine the data. | It is always a good idea to examine the data before jumping straight into calibration. Start by flagging data known to be bad from observing logs, then examine the data. | ||
At low frequencies there is a lot of RFI, some of it is very strong and persistent so lets get of rid of that first. Use plotms to make a plot of the XY and YX correlations on the secondary calibrator. These correlations should normally have little signal in them, certainly anything stronger than the calibrator flux will be interference. Specify axes of 'channel' and 'amp', zoom in and use the resulting plots to read off the worst channel ranges. To cut down on the amount of data we can use antenna selection "0&4", the longest baseline we're likely to use, if it is bad, the shorter ones will be worse. The following | At low frequencies there is a lot of RFI, some of it is very strong and persistent so lets get of rid of that first. Use plotms to make a plot of the XY and YX correlations on the secondary calibrator. These correlations should normally have little signal in them, certainly anything stronger than the calibrator flux will be interference. Specify axes of 'channel' and 'amp', zoom in and use the resulting plots to read off the worst channel ranges. To cut down on the amount of data we can use antenna selection "0&4", the longest baseline we're likely to use, if it is bad, the shorter ones will be worse. The following command flags the data in all the bad channel ranges we found, you may decide to excise a few more or fewer channel ranges. At the highest channel number (lowest frequencies) the RFI gets so bad, we've deleted large channel ranges. If you were desperate for spectral information in this range you could do a more careful job. Note that it is not necessary to be very accurate in this step, as we will do further flagging after this first pass. | ||
By default '''{{flagdata}}''' will save a copy of the existing set of flags ''before'' entering any new flags. This is so that you can easily revert any flags you have applied using the {{flagmanager}} task. This comes in very handy for those occasions where you accidentally flag more than you intended. | By default '''{{flagdata}}''' will save a copy of the existing set of flags ''before'' entering any new flags. This is so that you can easily revert any flags you have applied using the {{flagmanager}} task. This comes in very handy for those occasions where you accidentally flag more than you intended. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
flagdata(vis=msname0,mode='manual',spw='0:243~247;548~551;631~634;895~899;1012~1025;1205~1234;1421~1439;1495~1599;1614~1625;1675~1676;1815~2048') | |||
</source> | </source> | ||
Next we will do some clipping on the cross correlations | Next we will do some clipping on the cross correlations. To choose the clip level we checked the amplitudes on a XY and YX plot with all the data, to make sure we're not clipping any good data. After clipping we extend the flagged points using 'growaround=True' and also flag the corresponding parallel hand polarizations. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
flagdata(vis=msname0,mode='clip',clipminmax=[0, | flagdata(vis=msname0,mode='clip',clipminmax=[0,5],correlation='ABS,XY,YX') | ||
flagdata(vis=msname0,mode='extend',extendpols=True,growaround=True) | flagdata(vis=msname0,mode='extend',extendpols=True,growaround=True) | ||
</source> | </source> | ||
| Line 236: | Line 235: | ||
</source> | </source> | ||
After these steps the data is much improved and | After these steps the data is much improved but after replotting the secondary calibrator we notice one more flaw: there is a single integration with excess flux on the secondary calibrator. | ||
<source lang="python"> | |||
# In CASA | |||
flagdata(vis=msname0,mode='manual',timerange='14:15:30~14:15:40') | |||
</source> | |||
After fixing that we inspect the rest of the data and it is looking pretty clean. We could do some more flagging but it is probably not warranted unless we notice problems later in the reduction. | |||
<source lang="python"> | <source lang="python"> | ||
Revision as of 06:17, 28 July 2016
This CASA Guide is for Version 4.6 of CASA. If you are using a later version of CASA and this is the most recent available guide, then you should be able to use most, if not all, of this casaguide, as we try to limit script breaking changes in CASA development.
Overview
This CASA guide describes the loading, flagging, calibration and imaging of a two-pointing continuum data set taken with the Australia Telescope Compact Array (ATCA) of the nearby radio galaxy NGC612 NGC612. It has been adapted from the 3C391 Continuum Tutorial (CASA 4.6) for the VLA . The data were taken with 2048 MHz of bandwidth in 1 MHz channels, centered at 2.1 GHz, and recorded all polarizations. For ATCA data reduction there are two paths you can follow:
- If you are already familiar with the Miriad package, you can do the loading and optionally the flagging and calibration of the data in Miriad and then import the data into a CASA MeasurementSet with importmiriad.
- Alternatively, you can use importatca to load the ATCA archive files directly into CASA.
If this is your first attempt at using CASA for Compact Array data you are encouraged to start with a flagged and calibrated Miriad dataset and then try imaging the data following [this guide]. If you feel you are ready to tackle the full reduction job in CASA keep following this guide.
Obtaining the Data
To download the data needed for this tutorial you will need to use the [Australia Telescope Online Archive]. If you don't have an account already, you can request one [here]. Once logged in enter the project code C2728, click search and select the files 2012-10-25_0707.C2728, 2012-10-25_0903.C2728, 2012-10-25_1304.C2728 and 2012-10-25_1705.C2728 from the results. You can then use the download button to retrieve a tar file with the data, or get a file with the links to the data to download from another machine. (If that all seems like too much trouble, a ready made MS with the data is available for download [here](3.5GB download), you can extract that instead and continue in the section #The Observation.)
Once the download is complete, extract the files (within a working directory in which you will then run CASA):
# In a Terminal:
tar xvf datafiles.tar
How to Use This CASA Guide
Here are a number of possible ways to run CASA, described in more detail in Getting Started in CASA. In brief, there are at least three different ways to run CASA:
- Interactively examining task inputs. In this mode, one types taskname to load the task, inp to examine the inputs, and go once those inputs have been set to your satisfaction. Allowed inputs are shown in blue and bad inputs are colored red. The input parameters themselves are changed one by one, e.g., selectdata=T. Screenshots of the inputs to various tasks used in the data reduction below are provided, to illustrate which parameters need to be set. More detailed help can be obtained on any task by typing help taskname. Once a task is run, the set of inputs are stored and can be retrieved via tget taskname; subsequent runs will overwrite the previous tget file.
- Pseudo-interactively via task function calls. In this case, all of the desired inputs to a task are provided at once on the CASA command line. This tutorial is made up of such calls, which were developed by looking at the inputs for each task and deciding what needed to be changed from default values. For task function calls, only parameters that you want to be different from their defaults need to be set.
- Non-interactively via a script. A series of task function calls can be combined together into a script, and run from within CASA via execfile('scriptname.py'). This and other CASA Tutorial Guides have been designed to be extracted into a script via the script extractor by using the method described at the Extracting_scripts_from_these_tutorials page. Should you use the script generated by the script extractor for this CASA Guide, be aware that it will require some small amount of interaction related to the plotting, occasionally suggesting that you close the graphics window and hitting return in the terminal to proceed. It is in fact unnecessary to close the graphics windows (it is suggested that you do so purely to keep your desktop uncluttered).
If you are a relative novice or just new to CASA, it is strongly recommended to work through this tutorial by cutting and pasting the task function calls provided below after you have read all the associated explanations. Work at your own pace, look at the inputs to the tasks to see what other options exist, and read the help files. Later, when you are more comfortable, you might try to extract the script, modify it for your purposes, and begin to reduce other data.
Importing the ATCA archive data
Before beginning our data reduction, we must start CASA. If you have not used CASA before, some helpful tips are available on the Getting Started in CASA page.
Once you have CASA up and running in the directory containing the data, then start your data reduction by getting the data into a MeasurementSet. The task to do this, importatca, lets us choose which of the simultaneously recorded frequencies to load. In this case we just want the continuum data, which we can select with spw=0. There are two other frequency settings in this data, a 33 channel continuum band and a 2048 channel spectral zoom band centered on the HI emission from NGC612. We might use the latter for a future spectral line tutorial. We specify the options 'birdie' to remove known bad channels and 'noac' to discard the autocorrelations.
# In CASA
importatca(vis='ngc612.ms',files='*.C2728',spw=0,options='birdie,noac')
As the data loads a listing appears in the logger of the data encountered. The first part is reproduced here:
##########################################
##### Begin Task: importatca #####
importatca(files="*.C2728",vis="ngc612.ms.0",options="birdie,noac",spw=0,nscans=[0, 0],
lowfreq="0.1GHz",highfreq="999GHz",fields=[''],edge=8)
Expanded file names are :
/data/ngc612/2012-10-25_0707.C2728
/data/ngc612/2012-10-25_0903.C2728
/data/ngc612/2012-10-25_1304.C2728
/data/ngc612/2012-10-25_1705.C2728
Checking header of file /data/ngc612/2012-10-25_0707.C2728
CABB data detected
Reading file /data/ngc612/2012-10-25_0707.C2728
Scan # : 1
Object : 1934-638
Date : 2012/10/25/07:07:45
First data/header has NAnt=6, NChan=2049, NPol=4
Antenna : CA01 position:[-4.75168e+06, 2.79161e+06, -3.20048e+06]
Antenna : CA02 position:[-4.75159e+06, 2.79176e+06, -3.20048e+06]
Antenna : CA03 position:[-4.75156e+06, 2.79181e+06, -3.20048e+06]
Antenna : CA04 position:[-4.75135e+06, 2.79217e+06, -3.20048e+06]
Antenna : CA05 position:[-4.75129e+06, 2.79227e+06, -3.20048e+06]
Antenna : CA06 position:[-4.74939e+06, 2.79549e+06, -3.20048e+06]
IF 1 : Frequency = 2100 MHz, bandwidth = 2048MHz, #channels = 2049
: Polarizations XX - 9 , XY - 10 , YX - 11 , YY - 12
Scan 1 stored
Scan # : 2
Object : secndry_cal
Date : 2012/10/25/07:27:15
Scan 2 stored
Scan # : 3
Object : east_lobe
Date : 2012/10/25/07:31:15
Scan 3 stored
Scan # : 4
Object : secndry_cal
Date : 2012/10/25/07:32:25
Scan 4 stored
Scan # : 5
Object : east_lobe
Date : 2012/10/25/07:35:05
Scan 5 stored
Scan # : 6
Object : west_lobe
Date : 2012/10/25/07:40:45
Scan 6 stored
Scan # : 7
Object : east_lobe
Date : 2012/10/25/07:46:05
Scan 7 stored
Scan # : 8
Object : west_lobe
Date : 2012/10/25/07:51:25
Scan 8 stored
Scan # : 9
Object : east_lobe
Date : 2012/10/25/07:56:55
Scan 9 stored
Scan # : 10
Object : west_lobe
Date : 2012/10/25/08:02:15
Scan 10 stored
Scan # : 11
Object : east_lobe
Date : 2012/10/25/08:07:45
Scan 11 stored
Scan # : 12
Object : west_lobe
Date : 2012/10/25/08:13:05
Scan 12 stored
Scan # : 13
Object : east_lobe
Date : 2012/10/25/08:18:25
Scan 13 stored
Scan # : 14
Object : west_lobe
Date : 2012/10/25/08:23:45
Scan 14 stored
Scan # : 15
Object : secndry_cal
Date : 2012/10/25/08:29:05
Scan 15 stored
Scan # : 16
Object : secndry_cal
Date : 2012/10/25/08:31:45
Scan 16 stored
Scan # : 17
Object : east_lobe
Date : 2012/10/25/08:34:05
Scan 17 stored
Scan # : 18
Object : west_lobe
Date : 2012/10/25/08:39:45
Scan 18 stored
Scan # : 19
Object : secndry_cal
Date : 2012/10/25/08:45:05
Scan 19 stored
Scan # : 20
Object : east_lobe
Date : 2012/10/25/08:47:45
Scan 20 skipped
Scan # : 21
Object : 1934-638
Date : 2012/10/25/08:47:45
Scan 21 stored
Scan # : 22
Object : 1934-638
Date : 2012/10/25/08:54:05
IF 2 : Frequency = 1891 MHz, bandwidth = 2048MHz, #channels = 33
: Polarizations XX - 9 , XY - 10 , YX - 11 , YY - 12
End of File
Number of rows selected : 8910
Flagged : 5.1%
Antenna off source : 5.1%
ScanType (Point/Paddle): 0%
Scans that don't have valid or selected data will be marked 'skipped'. At the end a summary of the on-line flagging appears, we see about 5% of the data is flagged because antennas were not on source.
The Observation
The task listobs can be used to get a listing of the individual scans comprising the observation, the frequency setup, source list, and antenna locations. Since we've already seen the scans scroll past, we'll use verbose=False to get the summary:
# In CASA
listobs(vis='ngc612.ms',verbose=False)
From the source listing we see there are 5 sources, the primary calibrator 1934-638, a secondary calibrator, and two target sources called east_lobe and west_lobe and some data on 0537-441. We also see the data has a 2048 MHz bandwidth, with 1 MHz channels, centered on 2.1 GHz with 4 linear polarizations.
##### Begin Task: listobs #####
listobs(vis="ngc612.ms.0",selectdata=True,spw="",field="",antenna="",
uvrange="",timerange="",correlation="",scan="",intent="",
feed="",array="",observation="",verbose=False,listfile="",
listunfl=False,cachesize=50,overwrite=False)
================================================================================
MeasurementSet Name: /data/ngc612/casacal/ngc612.ms.0 MS Version 2
================================================================================
Observer: obs Project: C2728
Observation: ATCA(6 antennas)
Data records: 60465 Total elapsed time = 43730 seconds
Observed from 25-Oct-2012/07:07:39.9 to 25-Oct-2012/19:16:29.9 (UTC)
Compute subscan properties
Fields: 5
ID Code Name RA Decl Epoch SrcId nRows
0 C 1934-638 19:39:25.025995 -63.42.45.63000 J2000 0 6645
1 secndry_cal 00:51:09.502006 -42.26.33.29294 J2000 1 5880
2 east_lobe 01:34:16.760003 -36.30.13.52004 J2000 2 23160
3 west_lobe 01:33:34.630993 -36.29.21.54998 J2000 3 22515
4 C 0537-441 05:38:50.362001 -44.05.08.93995 J2000 4 2265
Spectral Windows: (1 unique spectral windows and 1 unique polarization setups)
SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) Corrs
0 2049 TOPO 3124.000 1000.000 2048000.0 2100.0000 XX XY YX YY
Antennas: 6 'name'='station'
ID= 0-3: 'CA01'='W98', 'CA02'='W109', 'CA03'='W113', 'CA04'='W140',
ID= 4-5: 'CA05'='W148', 'CA06'='W392'
##### End Task: listobs #####
##########################################
The antenna station names give away that this is a compact configuration, with CA01-CA05 close together and CA06 a long way away (multiply the station numbers by 15 to get distance in meters from station W0). It is actually a 750m array configuration, which means we would normally not include CA06 in the imaging. The galaxy ngc612 is quite large compared to the primary beam at 2 GHz, so two pointings were used to cover the emission with adequate sensitivity. The observation ran for close to 12 hours, this should give good uv coverage with the array in a short E-W configuration.
Examining and Editing the Data
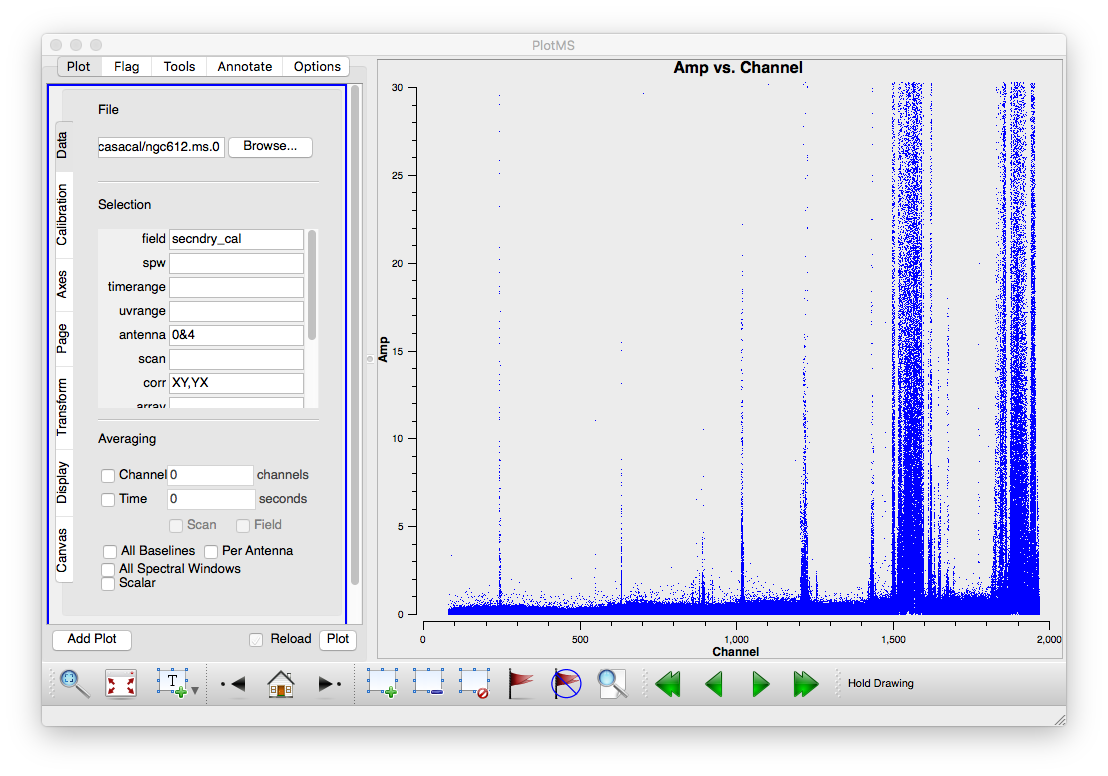
It is always a good idea to examine the data before jumping straight into calibration. Start by flagging data known to be bad from observing logs, then examine the data. At low frequencies there is a lot of RFI, some of it is very strong and persistent so lets get of rid of that first. Use plotms to make a plot of the XY and YX correlations on the secondary calibrator. These correlations should normally have little signal in them, certainly anything stronger than the calibrator flux will be interference. Specify axes of 'channel' and 'amp', zoom in and use the resulting plots to read off the worst channel ranges. To cut down on the amount of data we can use antenna selection "0&4", the longest baseline we're likely to use, if it is bad, the shorter ones will be worse. The following command flags the data in all the bad channel ranges we found, you may decide to excise a few more or fewer channel ranges. At the highest channel number (lowest frequencies) the RFI gets so bad, we've deleted large channel ranges. If you were desperate for spectral information in this range you could do a more careful job. Note that it is not necessary to be very accurate in this step, as we will do further flagging after this first pass.
By default flagdata will save a copy of the existing set of flags before entering any new flags. This is so that you can easily revert any flags you have applied using the flagmanager task. This comes in very handy for those occasions where you accidentally flag more than you intended.
# In CASA
flagdata(vis=msname0,mode='manual',spw='0:243~247;548~551;631~634;895~899;1012~1025;1205~1234;1421~1439;1495~1599;1614~1625;1675~1676;1815~2048')
Next we will do some clipping on the cross correlations. To choose the clip level we checked the amplitudes on a XY and YX plot with all the data, to make sure we're not clipping any good data. After clipping we extend the flagged points using 'growaround=True' and also flag the corresponding parallel hand polarizations.
# In CASA
flagdata(vis=msname0,mode='clip',clipminmax=[0,5],correlation='ABS,XY,YX')
flagdata(vis=msname0,mode='extend',extendpols=True,growaround=True)
As a final step we'll use the 'tfcrop' algorithm to find further bad data. It automatically does a flag extend to the other polarizations. Read the help for flagdata for the details on how these flagging algorithms work.
# In CASA
flagdata(vis=msname0,mode='tfcrop',usewindowstats='sum',correlation='ABS,XY,YX',halfwin=3)
After these steps the data is much improved but after replotting the secondary calibrator we notice one more flaw: there is a single integration with excess flux on the secondary calibrator.
# In CASA
flagdata(vis=msname0,mode='manual',timerange='14:15:30~14:15:40')
After fixing that we inspect the rest of the data and it is looking pretty clean. We could do some more flagging but it is probably not warranted unless we notice problems later in the reduction.
# In CASA
#split into 8 spw - need to use latest casa 4.7 - mstransform has bugs in 4.6
mstransform(vis=msname0,outputvis=msname,datacolumn='data',mode='channel',
regridms=True,spw='0',nspw=8)
#set flux scale for primary calibrator
setjy(vis=msname,field=pri,scalebychan=True,standard='Perley-Butler 2010')
#calibrate
# initial phase cal
gaincal(vis=msname,caltable=prefix+'.G0',field=pri,refant='CA03',gaintype='G',calmode='p',parang=T,solint='60s');
# bandpass
# snr dips below 3 for some channels
bandpass(vis=msname,caltable=prefix+'.B0',field=pri,spw='',refant='CA03',solnorm=True,solint='inf',bandtype='B',gaintable=[prefix+'.G0'],parang=T);
# solint='inf',bandtype='B',gaintable=[prefix+'.G0',prefix+'.K0'],spwmap=[[],[0,0,0,0,0,0,0,0]],parang=T);
# gain cal
# solution on secondary has SNR < 1 for some times/ants and gets flagged
gaincal(vis=msname,caltable=prefix+'.G1',field=pri+','+sec,refant='CA05',spw='*',gaintype='G',calmode='ap',parang=T,solint='60s',gaintable=[prefix+'.B0'])
polcal(vis=msname,caltable=prefix+'.D0',field=pri,refant='CA03',gaintable=[prefix+'.B0',prefix+'.G1'],poltype='Dlls',solint='inf')
# Get Q/U from gains, X feed is at 45 degrees (unless 7mm -> 135 deg)
qu = qufromgain(prefix+'.G1',fieldids=[0],paoffset=45.)
smodel=[1,qu[0],qu[1],0]
# Redo gaincal
bandpass(vis=msname,caltable=prefix+'.B1',field=pri,spw='',refant='CA03',solnorm=True,
solint='inf',bandtype='B',gaintable=[prefix+'.G1',prefix+'.D0'],parang=T);
gaincal(vis=msname,caltable=prefix+'.G2',field=pri,refant='CA03',spw='*',
gaintype='G',calmode='ap',parang=T,solint='int',gaintable=[prefix+'.B1',prefix+'.D0'])
gaincal(vis=msname,caltable=prefix+'.G2',field=sec,refant='CA03',spw='*',
gaintype='G',calmode='ap',parang=T,solint='int',gaintable=[prefix+'.B1',prefix+'.D0'],smodel=smodel,append=True)
polcal(vis=msname,caltable=prefix+'.D1',field=pri,refant='CA03',gaintable=[prefix+'.B1',prefix+'.G2'],poltype='Dlls',solint='inf')
# flux cal
fluxscale(vis=msname,caltable=prefix+'.G2',fluxtable=prefix+'.F0',reference='1934-638');
# Apply calibration
applycal(vis=msname,gaintable=[prefix+'.B1',prefix+'.D1',prefix+'.F0'],gainfield=[pri,pri,sec],parang=T)
# make first image
os.system('rm -rf tmp-ngc612*')
clean(vis='ngc612.ms',imagename='tmp-ngc612',spw='0,1,2:0~170',imagermode='mosaic',multiscale=[0,10,30],imsize=500,cell='5.0arcsec',stokes='IV',weighting='briggs',robust=0.5,antenna='!5',field='*_lobe')
# use viewer to create cleanbox file
# clean deeper image
#!rm -rf ngc612h.*
clean(vis='ngc612.ms',imagename='ngc612h1',spw='0,1,2:0~170',imagermode='mosaic',multiscale=[0,8,24],imsize=500,cell='5.0arcsec',stokes='IQUV',weighting='briggs',robust=0.5,niter=5000,threshold='0.5mJy',antenna='!5',mask='box [[01:34:28.51334, -036.32.55.9841], [01:33:14.14565, -036.25.15.8659]]',field='*_lobe')