IRAS16293 Band9 - Imaging for CASA 3.4: Difference between revisions
| Line 8: | Line 8: | ||
[http://almascience.nao.ac.jp/almadata/sciver/IRAS16293B9 East Asia] | [http://almascience.nao.ac.jp/almadata/sciver/IRAS16293B9 East Asia] | ||
This casa guide covers imaging of both continuum and spectral lines of IRAS16293 Band 9. The commands in this guide should be executed in CASA 3.4.0. Among other things, CASA 3.4.0 contains an improved antenna primary beam model for ALMA. In this casa guide you will also need the Analysis Utils package, so make sure you have it imported into CASA. | This casa guide covers imaging of both continuum and spectral lines of IRAS16293 Band 9. The commands in this guide should be executed in CASA 3.4.0. Among other things, CASA 3.4.0 contains an improved antenna primary beam model for ALMA. In this casa guide you will also need the [http://casaguides.nrao.edu/index.php?title=Analysis_Utilities Analysis Utils] package, so make sure you have it imported into CASA. | ||
Before we start with the imaging itself, do a {{listobs}} to the calibrated dataset, so you can check that everything is ok. | Before we start with the imaging itself, do a {{listobs}} to the calibrated dataset, so you can check that everything is ok. | ||
Revision as of 16:11, 31 May 2012
Overview
This section of the casa guide continues from IRAS16293_Band9_-_Calibration_for_CASA_3.4 . If you completed the calibration section, then you already have the final calibrated dataset. If you just started to work on this IRAS16293 B9 data, you can download the fully calibrated dataset IRAS16293_Band9_CalibratedMS from
This casa guide covers imaging of both continuum and spectral lines of IRAS16293 Band 9. The commands in this guide should be executed in CASA 3.4.0. Among other things, CASA 3.4.0 contains an improved antenna primary beam model for ALMA. In this casa guide you will also need the Analysis Utils package, so make sure you have it imported into CASA.
Before we start with the imaging itself, do a listobs to the calibrated dataset, so you can check that everything is ok.
# listobs to check data
listobs(vis='IRAS16293_Band9.rebin.ms',
listfile='IRAS16293_Band9.rebin.ms.listobs',verbose=F)
You should see the next information in the .listobs file.
Fields: 7 ID Code Name RA Decl Epoch SrcId nRows 0 none IRAS16293-2422-a 16:32:22.99200 -24.28.36.0000 J2000 0 7811 1 none IRAS16293-2422-a 16:32:22.47925 -24.28.36.0000 J2000 0 7829 2 none IRAS16293-2422-a 16:32:22.73563 -24.28.36.0000 J2000 0 7829 3 none IRAS16293-2422-a 16:32:22.73563 -24.28.32.5000 J2000 0 7829 4 none IRAS16293-2422-a 16:32:22.47925 -24.28.29.0000 J2000 0 7429 5 none IRAS16293-2422-a 16:32:22.73563 -24.28.29.0000 J2000 0 7151 6 none IRAS16293-2422-a 16:32:22.99200 -24.28.29.0000 J2000 0 7151 Spectral Windows: (4 unique spectral windows and 1 unique polarization setups) SpwID #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) Corrs 0 3840 TOPO 703312.744 488.28125 1875000 XX YY 1 3840 TOPO 692237.256 -488.28125 1875000 XX YY 2 3840 TOPO 690437.256 -488.28125 1875000 XX YY 3 3840 TOPO 688437.256 -488.28125 1875000 XX YY Antennas: 13 'name'='station' ID= 0-3: 'DA41'='A003', 'DA43'='A075', 'DA47'='A026', 'DV02'='A077', ID= 4-8: 'DV03'='A137', 'DV07'='A076', 'DV09'='A046', 'DV10'='A071', ID= 9-13: 'DV12'='A011', 'DV13'='A072', 'DV14'='A025', 'DV17'='A138',
Continuum Emission Imaging
As it is shown in Figure 2 of IRAS16293Band9, these observations have a 7 pointing mosaic, which you can visualize with the next command (see also Figure 1).
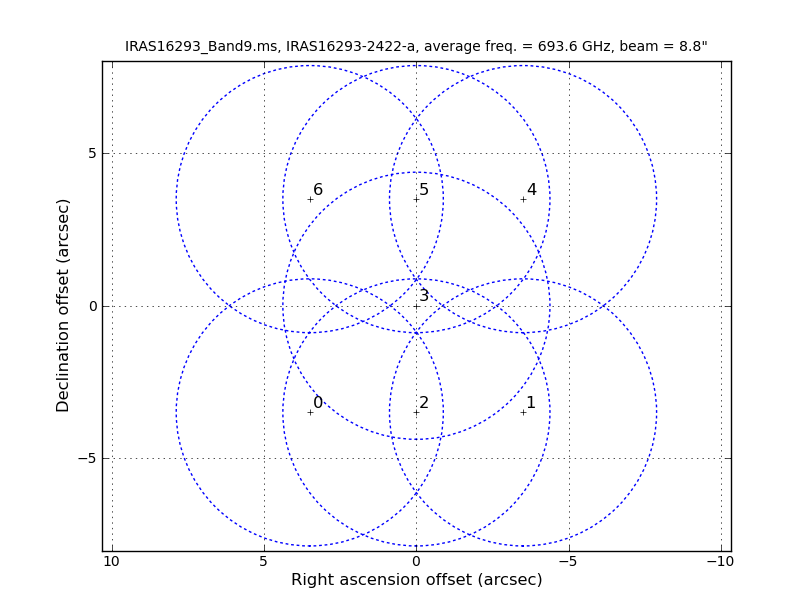
# Make a plot of the mosaic pattern with field ids identified
aU.plotMosaic(vis='IRAS16293_Band9.rebin.ms',sourceid='0',
doplot=True,figfile='mosaic_pattern.png')
You might wonder what of these pointings or fields have stronger line emission. You can know this by running the next command. We set spw='1' containing CO (6-5), and you can check all the plots for all the fields by just hitting the arrow for "next". Also see Figure 2 for an example of the output.
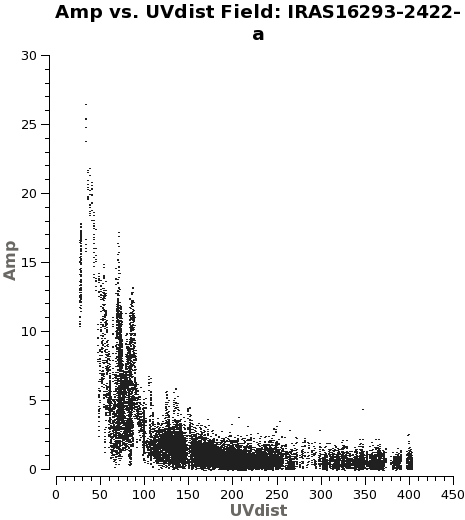
# See which mosaic fields have the brightest line emission
plotms(vis='IRAS16293_Band9.rebin.ms',
field='',xaxis='uvdist', yaxis='amp',
spw='1', avgchannel='3840',coloraxis='field',
iteraxis='field')
Another way to visualize the uv-amp distribution is with the next plotms. This will load all the fields or pointing into the plot, and you can change interactively between the spws using the appropriate button in plotms. See Figure 3 for a plot of spw 0.
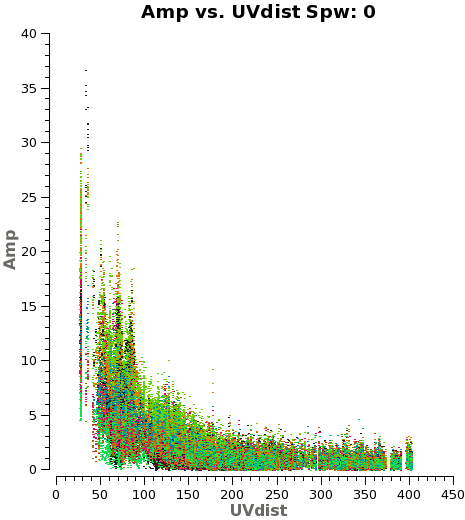
# Iterate over spws all fields
plotms(vis='IRAS16293_Band9.rebin.ms',
field='',xaxis='uvdist', yaxis='amp',
spw='', avgchannel='3840',coloraxis='field',
iteraxis='spw')
We need to note the channels that do not present line emission. To identify them we can plot the amplitude vs frequency for all the spectral windows. Figure 4 shows the output from the next command for spw 0 (field 3).
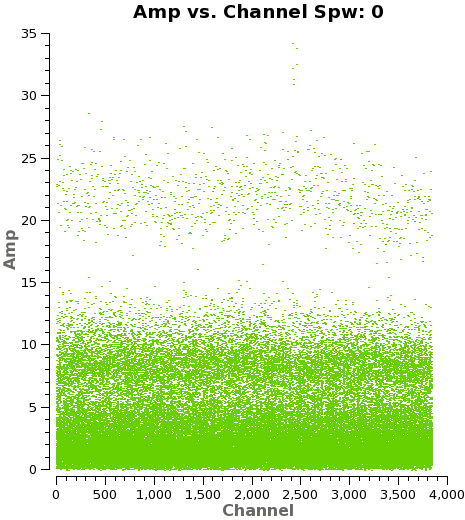
# Select Channels for uvcontsub. Average every 8 channels for
# increased S/N.
plotms(vis='IRAS16293_Band9.rebin.ms',
xaxis='channel', yaxis='amp',field='3',
spw='', avgtime='1e8',avgscan=T,coloraxis='field',
avgchannel='8',iteraxis='spw',ydatacolumn='corrected',
xselfscale=True,yselfscale=True)
Using the plots from the previous command you can select the line emission free channels just by looking for strong lines. To do this in the rigorous way, you would need to make dirty images and then select the continuum channels. Here we provide you with the channel selection we used for continuum emission.
The next flagmanager command will save the flags state in case you might need to re-do the steps from this point.
flagmanager(vis='IRAS16293_Band9.rebin.ms',mode='save',versionname='Original')
Now we will flag the channels identified to have line emission, so we will be ready to proceed with the continuum imaging.
# flagging the line channels
flagdata(vis='IRAS16293_Band9.rebin.ms',
spw='0:1700~2500,1:400~500;1100~1900;3000~3450,2:1700~2200,3100~3840,3:0~800;1600~2900;3300~3840',flagbackup=F)
To help clean to be faster, you can average 200 channels, as you do not need to have a high spectral resolution for continuum imaging.
split(vis='IRAS16293_Band9.rebin.ms',
outputvis='IRAS16293_Band9_CONT.msAVG',field='',
datacolumn='data',width=200,spw='0~3:20~3819')
Clean and self-cal the continuum
Now we proceed with the cleaning itself. For the clean task you will need to choose the right cell size, map size, etc. Below are the parameters that were used in this casaguide. Note that the command will be interactive and you will have to set the cleaning boxes.
# cleaning 1
os.system('rm -rf *CONTIN*')
clean(vis='IRAS16293_Band9_CONT.msAVG',
imagename='IRAS16293_Band9.CONTIN',
spw='0~3',field='',phasecenter='3',
mode='mfs',imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=T,
weighting='briggs',robust=0.5,
niter=10000, threshold='0.3mJy',usescratch=F)
It is important to check if all fields have some emission in the model data column, if not, or very weak you would want to exclude from the gaincal below. Because the bright Source B is in all the sidelobes all fields, do have some model data components. In Figure 5 you can see an example for the output from next command.
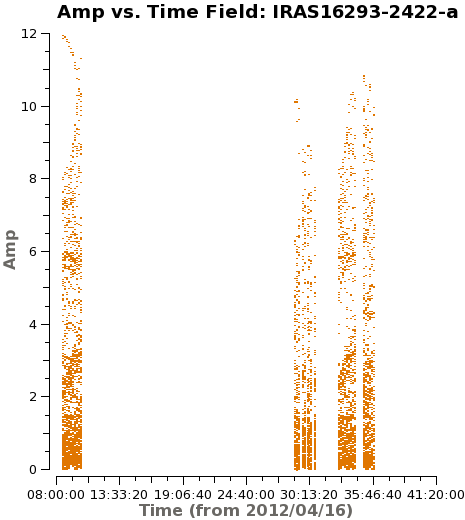
# checking the model data for all the fields
plotms(vis='IRAS16293_Band9_CONT.msAVG',
xaxis='time', yaxis='amp',field='',iteraxis='field',
spw='', avgchannel='4000',coloraxis='field',
ydatacolumn='model',xselfscale=True,yselfscale=True)
Now we proceed with gaincal to perform self calibration (for phases) on the data. Even with minsnr=3.0 there will be failed solutions.
# get one scan per field
os.system('rm -rf cont_pcal1')
gaincal(vis='IRAS16293_Band9_CONT.msAVG',caltable='cont_pcal1',gaintype='T',
refant='DV14',calmode='p',combine='',spw='',field='',
solint='inf',minsnr=3.0,minblperant=4)
The solutions for five antennas are shown in Figure 6, and are produced with the next plotcal. Be sure to check all the antennas.
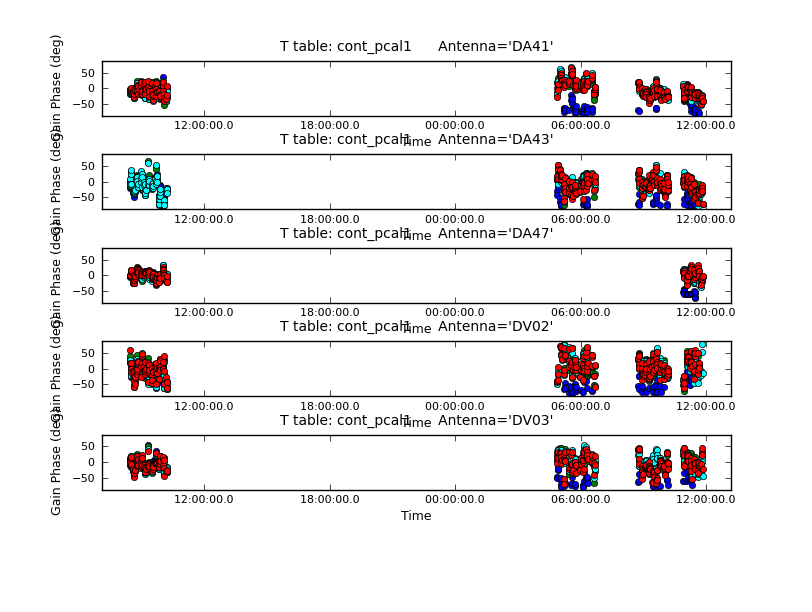
plotcal(caltable='cont_pcal1',xaxis='time',yaxis='phase',
spw='',iteration='antenna',subplot=511,plotrange=[0,0,-80,80],
antenna='',timerange='')
# The phases are remarkably stable for Band 9!
We then apply the table to the data to proceed right after with the proper cleaning, to check the improvement of the data.
# application of the table
applycal(vis='IRAS16293_Band9_CONT.msAVG',spwmap=[],spw='',
gaintable=['cont_pcal1'],calwt=F,flagbackup=F)
Now you are ready for the second cleaning. For this one we are using the mask resulting from the previous cleaning. In the next blocks of commands we will repeat this process (clean, viewer, gaincal, plotcal, applycal, clean, etc) to improve the overall quality of the image.
# cleaning 2
os.system('rm -rf *CONTIN_P1*')
clean(vis='IRAS16293_Band9_CONT.msAVG',
imagename='IRAS16293_Band9.CONTIN_P1',
spw='0~3',field='',phasecenter='3',
mode='mfs',imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=T,
weighting='briggs',robust=0.5,
mask='IRAS16293_Band9.CONTIN.mask',
niter=10000, threshold='0.3mJy')
# visualize the image
imview (raster=[{'file':'IRAS16293_Band9.CONTIN.image',
'range': [-0.01,0.5]},
{'file': 'IRAS16293_Band9.CONTIN_P1.image',
'range': [-0.01,0.5]}])
Second gaincal step for self calibration.
# second self-cal
os.system('rm -rf cont_pcal2')
gaincal(vis='IRAS16293_Band9_CONT.msAVG',caltable='cont_pcal2',gaintype='T',
refant='DV14',calmode='p',combine='',spw='',field='',
solint='inf',minsnr=3.0,minblperant=4)
#Checking the table
plotcal(caltable='cont_pcal2',xaxis='time',yaxis='phase',
spw='',iteration='antenna',subplot=511,plotrange=[0,0,-80,80],
antenna='',timerange='')
# Applying results to the data
applycal(vis='IRAS16293_Band9_CONT.msAVG',spwmap=[],spw='',
gaintable=['cont_pcal2'],calwt=F,flagbackup=F)
We continue with the next cleaning.
# Cleaning 3
os.system('rm -rf *CONTIN_P2*')
clean(vis='IRAS16293_Band9_CONT.msAVG',
imagename='IRAS16293_Band9.CONTIN_P2',
spw='0~3',field='',phasecenter='3',
mode='mfs',imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=T,
weighting='briggs',robust=0.5,
mask='IRAS16293_Band9.CONTIN.mask',
niter=10000, threshold='0.3mJy')
# to view the resulting image
imview (raster=[{'file':'IRAS16293_Band9.CONTIN_P1.image',
'range': [-0.01,0.5]},
{'file': 'IRAS16293_Band9.CONTIN_P2.image',
'range': [-0.01,0.5]}])
# There is very little change, the bold could try another iteration using a shorter
# solint phase self-cal. For now, moving on to amplitude.
Next iteration with gaincal, now using amplitude and phase for "calmode".
# For amplitude self-cal we only want to get one solution per spw, per dataset
os.system('rm -rf cont_apcal')
gaincal(vis='IRAS16293_Band9_CONT.msAVG',caltable='cont_apcal',gaintype='T',
refant='DV14',calmode='ap',combine='scan,field',spw='',field='',
solint='8000s',minsnr=3.0,minblperant=4,gaintable='cont_pcal2')
# ploting the table
plotcal(caltable='cont_apcal',xaxis='time',yaxis='amp',
spw='',iteration='antenna',subplot=511,plotrange=[0,0,0.5,2],
antenna='',timerange='')
# and applying to the data
applycal(vis='IRAS16293_Band9_CONT.msAVG',spwmap=[],spw='',
gaintable=['cont_pcal2','cont_apcal'],calwt=F,flagbackup=F)
# A large correction is present for the 2nd dataset. This amplitude
# error was causing many of the artifacts noticeable in the previous
# images.
Continuing the final cleaning step.
# Cleaning 4
os.system('rm -rf *CONTIN_AP*')
clean(vis='IRAS16293_Band9_CONT.msAVG',
imagename='IRAS16293_Band9.CONTIN_AP',
spw='0~3',field='',phasecenter='3',
mode='mfs',imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=T,
weighting='briggs',robust=0.5,
mask='IRAS16293_Band9.CONTIN_P2.mask',
niter=10000, threshold='0.3mJy')
# and to view the image
imview (raster=[{'file':'IRAS16293_Band9.CONTIN_P2.image',
'range': [-0.01,0.5]},
{'file': 'IRAS16293_Band9.CONTIN_AP.image',
'range': [-0.01,0.5]}])
We use now impbcor to produce an image corrected by the primary beam. We are outputting a sub-region of the cube because the area covered is large compared to the area of interest, and we want to keep the file size of the result as small as possible.
# correcting for primary beam
impbcor(imagename='IRAS16293_Band9.CONTIN_AP.image',
pbimage='IRAS16293_Band9.CONTIN_AP.flux',
outfile='IRAS16293_Band9.CONTIN_AP.image.pbcor.subim',
box='120,104,315,299')
# and checking the image
imview (raster=[{'file':'IRAS16293_Band9.CONTIN_AP.image',
'range': [-0.01,0.5]},
{'file': 'IRAS16293_Band9.CONTIN_AP.image.pbcor.subim',
'range': [-0.01,0.5]}])
In Figure 7 we show the final image of all this process. The rms noise measured is about 18 mJy/beam, the restoring beam resulted in 0.236864 x 0.171968 arcsec, PA=104.419 deg. The peak values for the two components are 2.6 and 0.7 Jy, while its fluxes are 10 and 6.8 Jy.
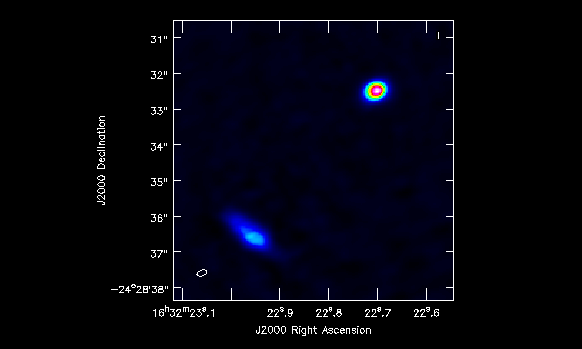
Now you can export your final image in fits format, so you can analyze it anytime.
exportfits(imagename='IRAS16293_Band9.CONTIN_AP.image.pbcor.subim',
fitsimage='IRAS16293_Band9.CONTIN_AP.image.pbcor.subim.fits')
Spectral Line Imaging
# Restore the flags made for the continuum imaging
flagmanager(vis='IRAS16293_Band9.rebin.ms',mode='restore',versionname='Original')
Then we need to subtract the continuum, using the line free channels
# Subtract the continuum from the line data
contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300'
uvcontsub(vis='IRAS16293_Band9.rebin.ms',fitspw=contspw,fitorder=1,combine='spw')
Before we commence with the cleaning we also need to apply the tables we produced in the self calibration steps.
# Apply final self-calibration to the line data
applycal(vis='IRAS16293_Band9.rebin.ms.contsub',spwmap=[],spw='',
gaintable=['cont_pcal2','cont_apcal'],calwt=F,flagbackup=F)
To make your work more effective we recommend you to clean cubes 1/2 spw at a time to make the image cubes a manageable size. Also, average every 2 channels since this is the native velocity resolution.
Some notes for this sections: the threshold was chosen after doing a couple of smaller test images. This is about 3 sigma in channels with weak line emission. The mask was also chosen from a few test images. With this set, there is no need to do an interactive cleaning process.
SPW 3
Cleaning the first half of the spw.
os.system('rm -rf *spw3a*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw3a',
spw='3',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=20,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
Second half.
os.system('rm -rf *spw3b*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw3b',
spw='3',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=1918,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
SPW 2
First half.
os.system('rm -rf *spw2a*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw2a',
spw='2',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=20,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
Second half.
os.system('rm -rf *spw2b*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw2b',
spw='2',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=1918,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
SPW 1
First half. This part has the CO (6-5) line. Note that highly resolved out CO is not well cleaned by this simple mask.
os.system('rm -rf *spw1a*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw1a',
spw='1',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=20,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
Second half.
os.system('rm -rf *spw1b*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw1b',
spw='1',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=1918,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
SPW 0
First half.
os.system('rm -rf *spw0a*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw0a',
spw='0',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=20,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
Second half.
os.system('rm -rf *spw0b*')
clean(vis='IRAS16293_Band9.rebin.ms.contsub',
imagename='IRAS16293_Band9.ms.spw0b',
spw='0',field='',phasecenter='3',
mode='channel',outframe='LSRK',
nchan=950,start=1918,width=2,
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=F,
mask=['circle[[16h32m22.7s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.94s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggs',robust=0.5,
niter=200, threshold='220.0mJy',usescratch=T)
H13CN
In this section we provide information about the imaging for a particular molecular line, H13CN, together with its moment mapping.
Producing a velocity cube
Here we present you an example of how to image a particular line of interest with the correct velocity.
You can use http://www.splatalogue.net/ to find rest frequencies. Another good reference is the 450 micron CSO survey of Orion: http://iopscience.iop.org/0067-0049/132/2/281/
First, determine velocity range for H13CN 690.55207 GHz with the help of the next plotms
plotms(vis='IRAS16293_Band9.ms.contsub',
xaxis='velocity', yaxis='amp',field='',
spw='1', avgtime='1e8',avgscan=T,coloraxis='field',
transform=True,freqframe='LSRK',restfreq='690.55207GHz',
avgchannel='8',
ydatacolumn='corrected',plotfile='Vel_H13CN.png')
Now do an interactive clean, making clean boxes for the emission. Stop cleaning when noise like residuals remain. Number of iterations between showing interactive window can be controlled within the interactive viewer. We used about 400 iterations.
os.system('rm -rf IRAS16293_Band9.H13CN_8_7*')
clean(vis='IRAS16293_Band9.ms.contsub',
imagename='IRAS16293_Band9.H13CN_8_7',
spw='1',field='',phasecenter='3',
mode='velocity',outframe='LSRK',restfreq='690.55207GHz',
nchan=60,start='-10km/s',width='0.5km/s',
imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=T,
weighting='briggs',robust=0.5,
niter=2000, threshold='220.0mJy')
You need to correct for the primary beam attenuation and subimage the cubes to a smaller more manageable size since most of the mosaic is empty.
spw=[0,1,2,3]
for i in spw:
impbcor(imagename='IRAS16293_Band9.ms.spw'+str(i)+'a.image',
pbimage='IRAS16293_Band9.ms.spw'+str(i)+'a.flux',
outfile='IRAS16293_Band9.ms.spw'+str(i)+'a.image.pbcor.subim',
box='120,104,315,299')
spw=[0,1,2,3]
for i in spw:
impbcor(imagename='IRAS16293_Band9.ms.spw'+str(i)+'b.image',
pbimage='IRAS16293_Band9.ms.spw'+str(i)+'b.flux',
outfile='IRAS16293_Band9.ms.spw'+str(i)+'b.image.pbcor.subim',
box='120,104,315,299')
impbcor(imagename='IRAS16293_Band9.H13CN_8_7.image',
pbimage='IRAS16293_Band9.H13CN_8_7.flux',
outfile='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim',
box='120,104,315,299')
To enable analysis in other software packages, you can export the data from CASA to FITS format using exportfits
spw=[0,1,2,3]
for i in spw:
exportfits(imagename='IRAS16293_Band9.ms.spw'+str(i)+'a.image.pbcor.subim',
fitsimage='IRAS16293_Band9.ms.spw'+str(i)+'a.image.pbcor.subim.fits')
spw=[0,1,2,3]
for i in spw:
exportfits(imagename='IRAS16293_Band9.ms.spw'+str(i)+'b.image.pbcor.subim',
fitsimage='IRAS16293_Band9.ms.spw'+str(i)+'b.image.pbcor.subim.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.fits')
Moments
To proceed with the moment maps generation, you will need to determine the channel range and pixel range for inclusion for higher order moments.
viewer('IRAS16293_Band9.H13CN_8_7.image.pbcor.subim')
Generation of moment 0
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.mom0')
immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim',moments=[0],
outfile='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.mom0',
chans='10~44')
which you can see in Figure 8. To produce that figure run the following command
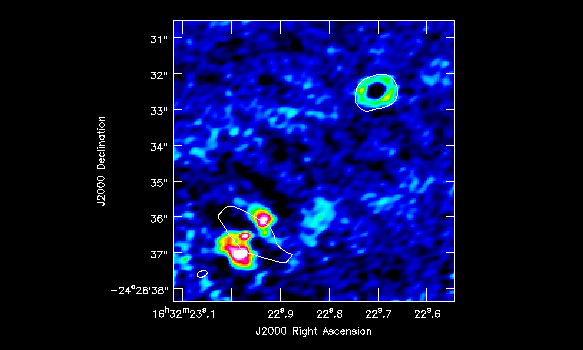
imview( raster=[ {'file':'IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.mom0',
'range': [-1.,4.8]} ],
contour={'file':'IRAS16293_Band9.CONTIN_AP.image.pbcor.subim',
'base':0, 'unit':0.025, 'levels':[3]},
out='IRAS16293_Band9.H13CN_8_7.mom0.png')
One cannot make higher order moments that simultaneously include both positive and negative emission. So we separate them below. Also, we need to restrict the higher order moments to regions of real signal. Below we demonstrate using the clean mask for this purpose. We must first subimage it to match the primary beam corrected image. We also set the included pixels to about 5sigma.
immath(imagename='IRAS16293_Band9.H13CN_8_7.mask',
mode='evalexpr',expr='IM0',box='120,104,315,299',
outfile='IRAS16293_Band9.H13CN_8_7.mask.subim')
Producing moment 1 and 2
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive*')
immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim',moments=[1,2],
outfile='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive',
mask='IRAS16293_Band9.H13CN_8_7.mask.subim',
chans='10~44',includepix=[0.5,100])
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative*')
immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim',moments=[1,2],
outfile='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative',
mask='IRAS16293_Band9.H13CN_8_7.mask.subim',
chans='10~44',includepix=[-100,-0.5])
Visualizing the maps
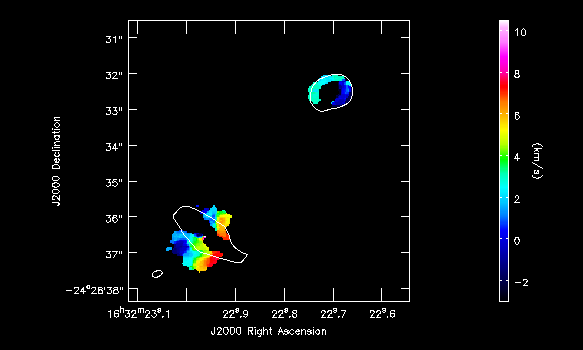
imview( raster=[ {'file':'IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative.weighted_coord',
'colorwedge':True}],
contour={'file':'IRAS16293_Band9.CONTIN_AP.image.pbcor.subim',
'base':0, 'unit':0.025, 'levels':[3]},
out='IRAS16293_Band9.H13CN_8_7.Negative_mom1.png')
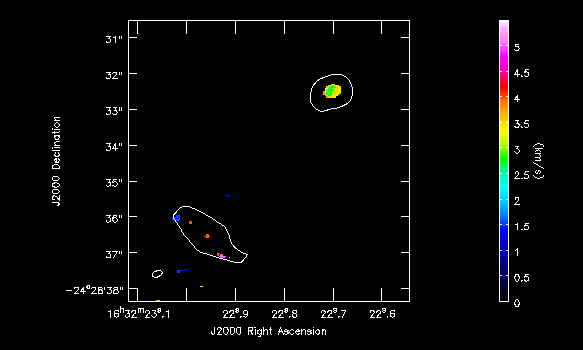
imview( raster=[ {'file':'IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive.weighted_coord',
'colorwedge':True}],
contour={'file':'IRAS16293_Band9.CONTIN_AP.image.pbcor.subim',
'base':0, 'unit':0.025, 'levels':[3]},
out='IRAS16293_Band9.H13CN_8_7.Positive_mom1.png')
And finally export the result to FITS format
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.mom0',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.mom0.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative.weighted_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative.weighted_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive.weighted_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive.weighted_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative.weighted_dispersion_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Negative.weighted_dispersion_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive.weighted_dispersion_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.pbcor.subim.Positive.weighted_dispersion_coord.fits')
Last checked on CASA Version 3.4.0.