IRAS16293 Band9 - Imaging for CASA 3.4: Difference between revisions
No edit summary |
No edit summary |
||
| Line 96: | Line 96: | ||
Using the plots from the previous command you can select the line emission free channels just by looking for strong lines. To do this in the rigorous way, you would need to make dirty images and then select the continuum channels. | Using the plots from the previous command you can select the line emission free channels just by looking for strong lines. To do this in the rigorous way, you would need to make dirty images and then select the continuum channels. | ||
Here we provide you with the channel selection we used for emission. | Here we provide you with the channel selection we used for continuum emission. | ||
Now {{uvcontsub}} will be used to calculate the level of continuum (based on your selection of continuum channels) and subtract that from the dataset. | Now {{uvcontsub}} will be used to calculate the level of continuum (based on your selection of continuum channels) and subtract that from the dataset. | ||
| Line 104: | Line 104: | ||
contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300' | contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300' | ||
uvcontsub(vis='IRAS16293_Band9.ms',fitspw=contspw,fitorder=1,combine='spw') | uvcontsub(vis='IRAS16293_Band9.ms',fitspw=contspw,fitorder=1,combine='spw') | ||
</source> | |||
The next {{flagmanager}} command will save the flags state in case you might need to re-do the continuum subtraction. | |||
<source lang="python"> | |||
flagmanager(vis='IRAS16293_Band9.ms',mode='save',versionname='Original') | |||
# NOTE: If uvcontsub needs to be run again restore flagged channels | |||
# first | |||
# flagmanager(vis='IRAS16293_Band9.ms',mode='restore',versionname='Original') | |||
</source> | |||
Now we will flag the channels we did not choose for continuum subtraction, ie the line emission channels. We will be left with only continuum channels. | |||
<source lang="python"> | |||
# flagging the line channels | |||
flagdata(vis='IRAS16293_Band9.ms', | |||
spw='0:1700~2500,1:400~500;1100~1900;3000~3450,2:1700~2200,3100~3840,3:0~800;1600~2900;3300~3840',flagbackup=F) | |||
</source> | |||
Note that more continuum imaging we do not need to have a high spectral resolution, so we will average 200 channels. This will help CASA imaging to run faster. | |||
<source lang="python"> | |||
# Make a channel averaged version for speedier continuum imaging | |||
split(vis='IRAS16293_Band9.ms', | |||
outputvis='IRAS16293_Band9_CONT.msAVG',field='', | |||
datacolumn='data',width=200,spw='0~3:20~3819') | |||
</source> | |||
Next you need to execute the next commands to fix some issues in the HISTORY section of the data. | |||
<source lang="python"> | |||
# Fix a bug with History table | |||
tb.open('IRAS16293_Band9_CONT.msAVG/HISTORY', nomodify = False) | |||
a = tb.rownumbers() | |||
tb.removerows(a) | |||
tb.close() | |||
tb.open('IRAS16293_Band9.ms.contsub/HISTORY', nomodify = False) | |||
a = tb.rownumbers() | |||
tb.removerows(a) | |||
tb.close() | |||
</source> | |||
=== Clean and self-cal the continuum === | |||
<source lang="python"> | |||
# cleaning the continuum | |||
os.system('rm -rf *CONTIN*') | |||
clean(vis='IRAS16293_Band9_CONT.msAVG', | |||
imagename='IRAS16293_Band9.CONTIN', | |||
spw='0~3',field='',phasecenter='3', | |||
mode='mfs',imagermode='mosaic', | |||
imsize=500,cell='0.04arcsec',minpb=0.25, | |||
interactive=T, | |||
weighting='briggs',robust=0.5, | |||
niter=10000, threshold='0.3mJy') | |||
</source> | </source> | ||
{{Checked 3.4.0}} | {{Checked 3.4.0}} | ||
Revision as of 18:32, 23 May 2012
Overview
This section of the casa guide continues from the last step of the calibration one. If you completed the calibration section, then you already have the final calibrated dataset. If you just started to work on this IRAS16293 B9 data, you can download the fully calibrated dataset IRAS16293_Band9_CalibratedMS from
This casa guide covers imaging of both continuum and line of IRAS16293 Band 9. The commands in this guide should be executed in CASA 3.4.0 since CASA 3.3.0 would be very slow for the cleaning process of a big dataset like this one for IRAS16293 Band 9. In this casa guide you will also need the Analysis Utils package, so make sure you have it imported into CASA.
Before we start with the imaging itself, do a listobs to the calibrated dataset, so you can check that everything is ok.
# listobs to check data
listobs(vis='IRAS16293_Band9.ms',
listfile='IRAS16293_Band9.ms.listobs',verbose=F)
You should see the next information in the .listobs file.
Fields: 7 ID Code Name RA Decl Epoch SrcId 0 none IRAS16293-2422-a 16:32:22.99200 -24.28.36.0000 J2000 0 1 none IRAS16293-2422-a 16:32:22.47925 -24.28.36.0000 J2000 0 2 none IRAS16293-2422-a 16:32:22.73563 -24.28.36.0000 J2000 0 3 none IRAS16293-2422-a 16:32:22.73563 -24.28.32.5000 J2000 0 4 none IRAS16293-2422-a 16:32:22.47925 -24.28.29.0000 J2000 0 5 none IRAS16293-2422-a 16:32:22.73563 -24.28.29.0000 J2000 0 6 none IRAS16293-2422-a 16:32:22.99200 -24.28.29.0000 J2000 0 (nVis = Total number of time/baseline visibilities per field) Spectral Windows: (4 unique spectral windows and 1 unique polarization setups) SpwID #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) Corrs 0 3840 TOPO 703312.744 488.28125 1875000 XX YY 1 3840 TOPO 692237.256 488.28125 1875000 XX YY 2 3840 TOPO 690437.256 488.28125 1875000 XX YY 3 3840 TOPO 688437.256 488.28125 1875000 XX YY Antennas: 14 'name'='station' ID= 0-3: 'DA41'='A003', 'DA43'='A075', 'DA47'='A026', 'DV02'='A077', ID= 4-7: 'DV03'='A137', 'DV05'='A082', 'DV07'='A076', 'DV09'='A046', ID= 8-11: 'DV10'='A071', 'DV12'='A011', 'DV13'='A072', 'DV14'='A025', ID= 12-13: 'DV17'='A138'
Continuum Emission Imaging
As it is shown in Figures 2 and 3 of IRAS16293Band9, these observations have a 7 pointing mosaic, which you can visualize with the next command (see also Figure 1).
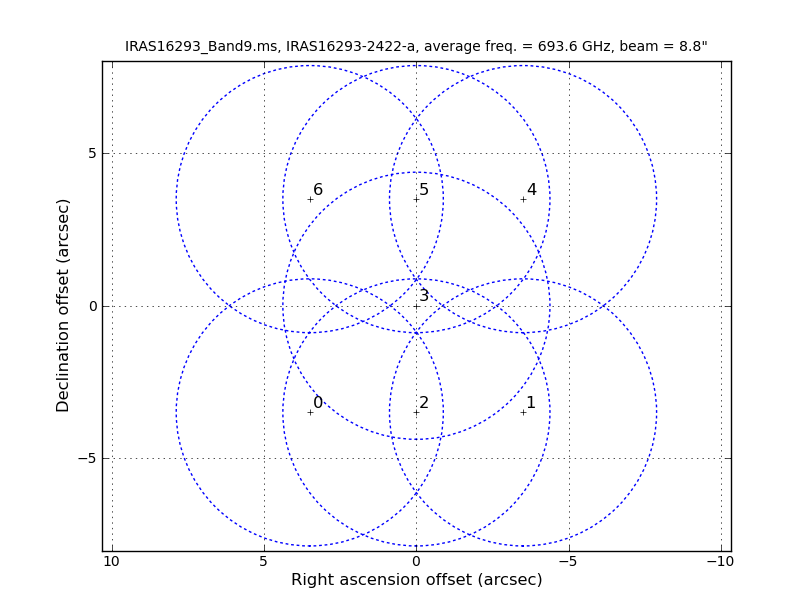
# Make a plot of the mosaic pattern with field ids identified
aU.plotMosaic(vis='IRAS16293_Band9.ms',sourceid='0',
doplot=True,figfile='mosaic_pattern.png')
You might wander what of these pointings or fields have stronger line emission. You can know this by running the next command. We set spw='1' containing CO (6-5), and you can check all the plots for all the fields by just hitting the arrow for "next". Also see Figure 2 for an example of the output.
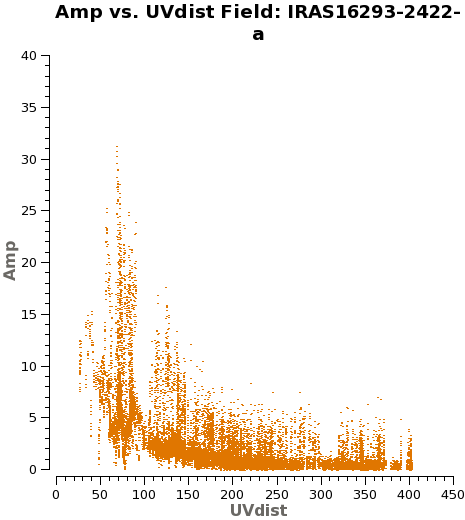
# See which mosaic fields have the brightest line emission
plotms(vis='IRAS16293_Band9.ms',
field='',xaxis='uvdist', yaxis='amp',
spw='1', avgchannel='3840',coloraxis='field',
iteraxis='field')
Another way to visualize the uv-amp distribution is with the next plotms. This will load all the fields or pointing into the plot, and you can change interactively between the spws using the appropriate button in plotms. See Figure 3 for a plot of spw 1.
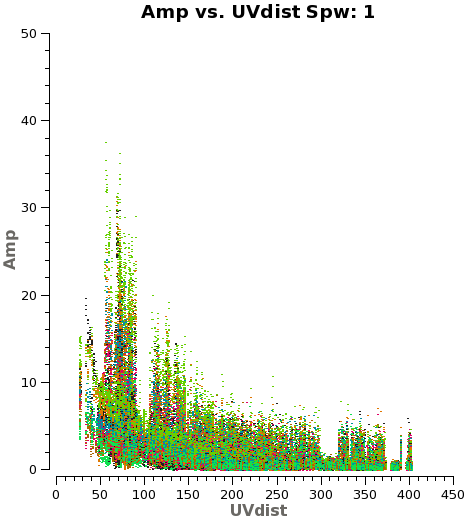
# Iterate over spws all fields
plotms(vis='IRAS16293_Band9.ms',
field='',xaxis='uvdist', yaxis='amp',
spw='', avgchannel='3840',coloraxis='field',
iteraxis='spw')
We need to note the channels that do not present line emission. To identify them we can plot the amplitude vs frequency for all the spectral windows. Figure 4 shows the output from the next command for spw 1.
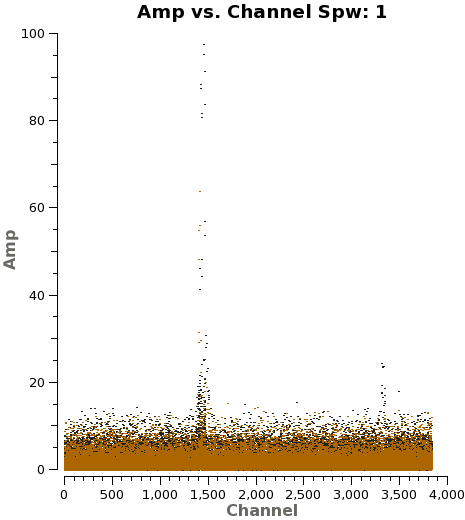
# Select Channels for uvcontsub. Average every 8 channels for
# increased S/N.
plotms(vis='IRAS16293_Band9.ms',
xaxis='channel', yaxis='amp',field='0,4',
spw='', avgtime='1e8',avgscan=T,coloraxis='field',
avgchannel='8',iteraxis='spw',ydatacolumn='corrected',
xselfscale=True,yselfscale=True)
Using the plots from the previous command you can select the line emission free channels just by looking for strong lines. To do this in the rigorous way, you would need to make dirty images and then select the continuum channels. Here we provide you with the channel selection we used for continuum emission.
Now uvcontsub will be used to calculate the level of continuum (based on your selection of continuum channels) and subtract that from the dataset.
# subtracting the continuum
contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300'
uvcontsub(vis='IRAS16293_Band9.ms',fitspw=contspw,fitorder=1,combine='spw')
The next flagmanager command will save the flags state in case you might need to re-do the continuum subtraction.
flagmanager(vis='IRAS16293_Band9.ms',mode='save',versionname='Original')
# NOTE: If uvcontsub needs to be run again restore flagged channels
# first
# flagmanager(vis='IRAS16293_Band9.ms',mode='restore',versionname='Original')
Now we will flag the channels we did not choose for continuum subtraction, ie the line emission channels. We will be left with only continuum channels.
# flagging the line channels
flagdata(vis='IRAS16293_Band9.ms',
spw='0:1700~2500,1:400~500;1100~1900;3000~3450,2:1700~2200,3100~3840,3:0~800;1600~2900;3300~3840',flagbackup=F)
Note that more continuum imaging we do not need to have a high spectral resolution, so we will average 200 channels. This will help CASA imaging to run faster.
# Make a channel averaged version for speedier continuum imaging
split(vis='IRAS16293_Band9.ms',
outputvis='IRAS16293_Band9_CONT.msAVG',field='',
datacolumn='data',width=200,spw='0~3:20~3819')
Next you need to execute the next commands to fix some issues in the HISTORY section of the data.
# Fix a bug with History table
tb.open('IRAS16293_Band9_CONT.msAVG/HISTORY', nomodify = False)
a = tb.rownumbers()
tb.removerows(a)
tb.close()
tb.open('IRAS16293_Band9.ms.contsub/HISTORY', nomodify = False)
a = tb.rownumbers()
tb.removerows(a)
tb.close()
Clean and self-cal the continuum
# cleaning the continuum
os.system('rm -rf *CONTIN*')
clean(vis='IRAS16293_Band9_CONT.msAVG',
imagename='IRAS16293_Band9.CONTIN',
spw='0~3',field='',phasecenter='3',
mode='mfs',imagermode='mosaic',
imsize=500,cell='0.04arcsec',minpb=0.25,
interactive=T,
weighting='briggs',robust=0.5,
niter=10000, threshold='0.3mJy')
Last checked on CASA Version 3.4.0.