Inspecting Data: Difference between revisions
No edit summary |
No edit summary |
||
| Line 22: | Line 22: | ||
Looking at this, we can see that our MS contains only a single object, 3C48, and a total of 12 scans of ~1.5-minute duration each with 10s integrations. There are 15 spectral windows, the first half containing RR and second half with LL polarizations. The "nRows" column tells us the number of rows for a particular scan in the MS. Each MS row has a unique timestamp, baseline, and SPW; for example, Scan 3 has 50544 / 16 (SPWs) / 351 (baselines) = 9 integrations, which makes sense for 10-s integrations and a 90-s scan length. | Looking at this, we can see that our MS contains only a single object, 3C48, and a total of 12 scans of ~1.5-minute duration each with 10s integrations. There are 15 spectral windows, the first half containing RR and second half with LL polarizations. The "nRows" column tells us the number of rows for a particular scan in the MS. Each MS row has a unique timestamp, baseline, and SPW; for example, Scan 3 has 50544 / 16 (SPWs) / 351 (baselines) = 9 integrations, which makes sense for 10-s integrations and a 90-s scan length. | ||
In order to get much of this information into a Python array, it is helpful to use the {{listsdm}} task. In order to get this to work, you will need the original SDM tables, which can be downloaded from the archive with your data (check "Include verbatim SDM tables in MS") or stand-alone ("SDM tables only (no visibiliites)" gets just the tables; ignore the file size, this will only be a few MB). | In order to get much of this information into a Python array, it is helpful to use the {{listsdm}} task. In order to get this to work, you will need the original SDM tables, which can be downloaded from the archive with your data (check "Include verbatim SDM tables in MS") or stand-alone ("SDM tables only (no visibiliites)" gets just the tables; ignore the file size, this will only be a few hundred MB). Then, you can type: | ||
<source lang='python'> | |||
# In CASA | |||
scanList = listsdm(vis='TVER0002.sb2568947.eb2579996.55518.22356400463.ms') | |||
</source> | |||
{{listsdm}} will output information both to the logger window and to a Python dictionary which we've called "scanList" -- this can be useful, for example, if you're scripting your processing, or if you have a large dataset (in which case {{listobs}} can be slow to execute). | |||
== Checking the online flags == | == Checking the online flags == | ||
Revision as of 21:29, 20 April 2012
Overview
Now that you've obtained your measurement set (MS), you will probably want to look at it carefully to see what it contains. This CASA Guide will describe a set of CASA tasks designed to help you evaluate and understand the data in an MS.
Following the example in Loading Data, we will be using the MS TVER0002.sb2568947.eb2579996.55518.22356400463.ms in this Guide. Please see Loading Data for instructions on acquiring this dataset, or use your own MS for practice.
Initial inspection: listobs
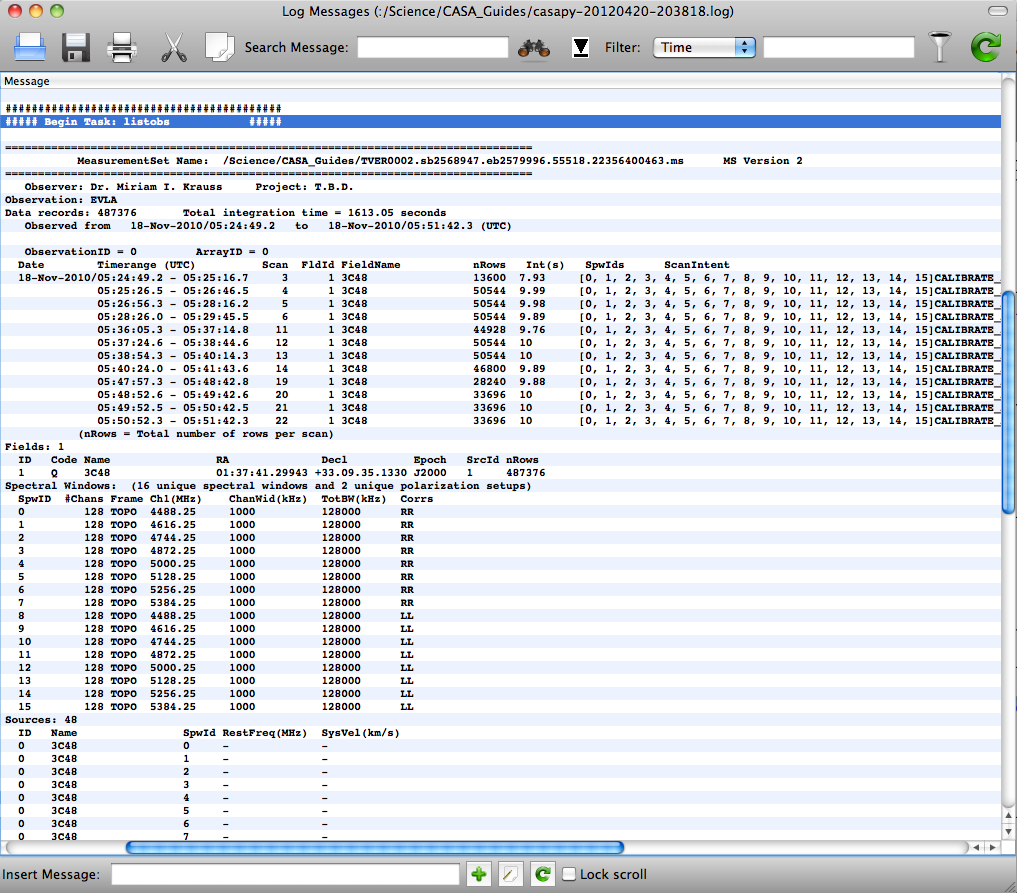
The best way to get a quick overview of the data in an MS is by using listobs:
# In CASA
listobs(vis='TVER0002.sb2568947.eb2579996.55518.22356400463.ms')
This sends useful information about the MS to the logger window, as well as to the current log file in the directory in which you started CASA.
Looking at this, we can see that our MS contains only a single object, 3C48, and a total of 12 scans of ~1.5-minute duration each with 10s integrations. There are 15 spectral windows, the first half containing RR and second half with LL polarizations. The "nRows" column tells us the number of rows for a particular scan in the MS. Each MS row has a unique timestamp, baseline, and SPW; for example, Scan 3 has 50544 / 16 (SPWs) / 351 (baselines) = 9 integrations, which makes sense for 10-s integrations and a 90-s scan length.
In order to get much of this information into a Python array, it is helpful to use the listsdm task. In order to get this to work, you will need the original SDM tables, which can be downloaded from the archive with your data (check "Include verbatim SDM tables in MS") or stand-alone ("SDM tables only (no visibiliites)" gets just the tables; ignore the file size, this will only be a few hundred MB). Then, you can type:
# In CASA
scanList = listsdm(vis='TVER0002.sb2568947.eb2579996.55518.22356400463.ms')
listsdm will output information both to the logger window and to a Python dictionary which we've called "scanList" -- this can be useful, for example, if you're scripting your processing, or if you have a large dataset (in which case listobs can be slow to execute).
Checking the online flags
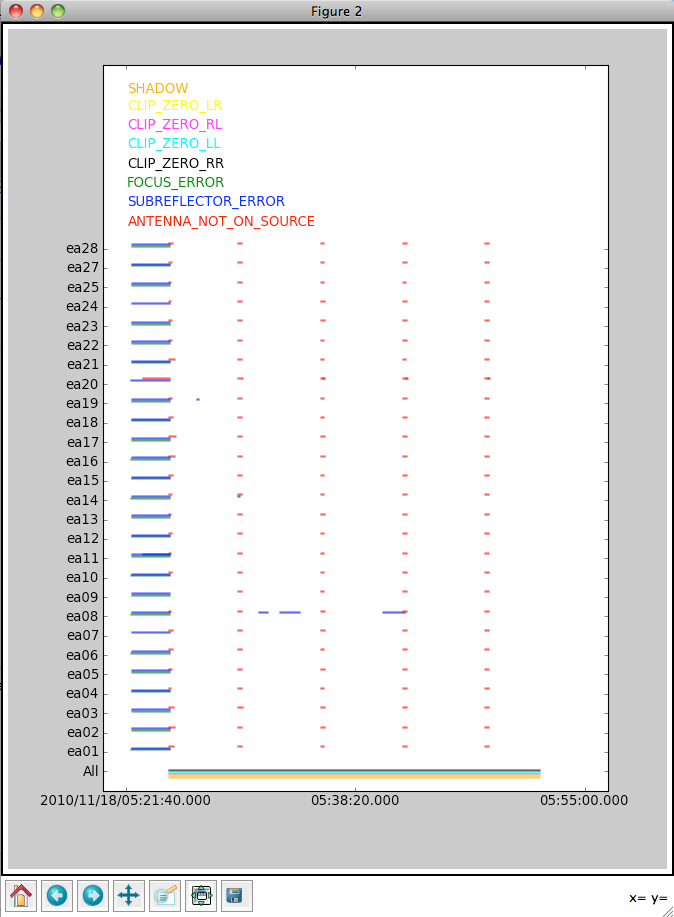
It's useful to start by checking which data have already been deleted (flagged) by the "online" flags (including time periods when the antennas were pointing off-source, or there were focus or subreflector errors), as well as data that were flagged because they were shadowed (blocked by a nearby antenna) or zero-valued (indicating a correlator error).
To do this, use the Template:Tflagcmd task to produce a plot:
# In CASA
tflagcmd(vis='TVER0002.sb2568947.eb2579996.55518.22356400463.ms',action='plot')
From this, we can see that antenna ea08 had some subreflector issues, and lengths of time when an antenna was not "on source" (i.e., it was slewing from one object to the next) varied a bit according to antenna. This accounts for the fact that the integration times provided by listobs are not exactly 10 s: the first integration of a given scan begins when the first antenna arrives on-source; since this does not happen simultaneously for all antennas, and the given integration time is an average across antennas, we can get values like 7.93 s (as for Scan 3).
Although the shadow and clip flags are plotted as well, the exact times affected by these flags are not known, so do not be alarmed by the fact that they appear to span the entire plot -- this is rarely actually the case.
Unfortunately, since we requested time-averaged data from the archive, the flagged data are not included in the MS. If you are concerned about the possible erroneous deletion of good data, it's best to download the complete MS, inspect it with plotms (see below), and then perform time and / or frequency averaging.
Checking the weather information
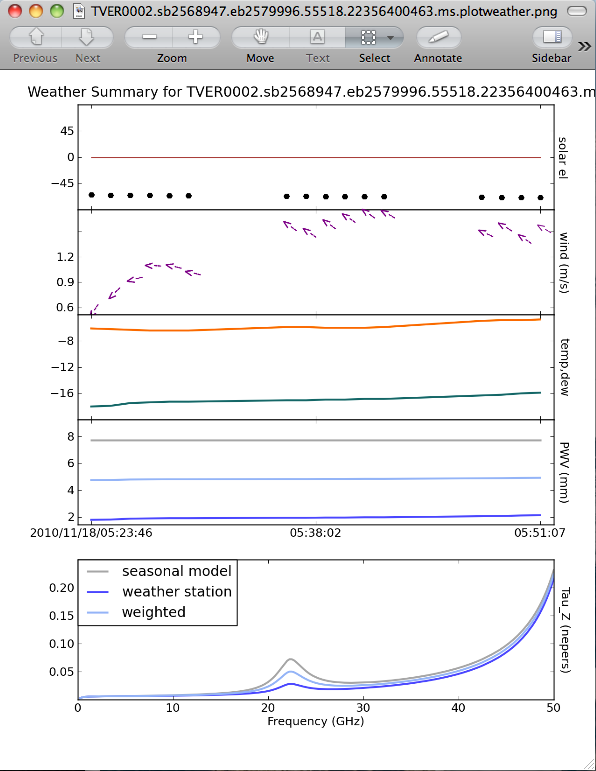
Another good thing to check is what the weather conditions were like during the observation. The task that does this is plotweather:
# In CASA
opacities = plotweather(vis='TVER0002.sb2568947.eb2579996.55518.22356400463.ms')
The plot is automatically named vis + ".plotweather.png," so in this case, TVER0002.sb2568947.eb2579996.55518.22356400463.ms.plotweather.png. Use your favorite graphics tool to open the file and inspect the information.
The top panel shows the solar elevation -- in this case, the observation was done at night (the black disk represents the sun, below the red line, which is the horizon). The second panel shows the wind speed and direction -- here, the wind was pretty calm (between 0.6 and 1.7 m/s). The temperature and dewpoint are shown in the next panel. This observation was performed in mid-November, and it was quite cold!
The last two panels plot the precipitable water vapor (PWV) and opacity (Tau_z) for three quantities: the value determined from the "seasonal model," the JVLA weather station, and a weighted average. See EVLA Memo 143 for more details.
In addition, you may notice that we set a variable opacities to capture output from plotweather. This task outputs a list of opacities, one for each spectral window, to use for calibration.