Image Continuum CASA 6.1.1: Difference between revisions
m Tashton moved page Image Continuum to Image Continuum CASA 6.1.1 |
|||
| (54 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
== Check CASA version == | == Check CASA version == | ||
This template is for use in CASA versions | This template is for use in CASA versions 6.1.1.15 and greater. The following code checks the version of CASA and exits if it is less than 6.1.1.15. You can download the appropriate version of CASA from [https://casa.nrao.edu/casa_obtaining.shtml Obtaining CASA] . See [[Guide_NA_ImagingTemplate#Prepare for Imaging | Prepare for Imaging ]] section on the main page of this guide for more information. | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
import re | import re | ||
try: | |||
import casalith | |||
except: | |||
print("Script requires CASA 6.0 or greater") | |||
if | if casalith.compare_version("<",[6,1,1,15]): | ||
print("Please use CASA version greater than or equal to 6.1.1-15 with this script") | |||
</source> | </source> | ||
== Create an Averaged Continuum MS == | == Create an Averaged Continuum MS == | ||
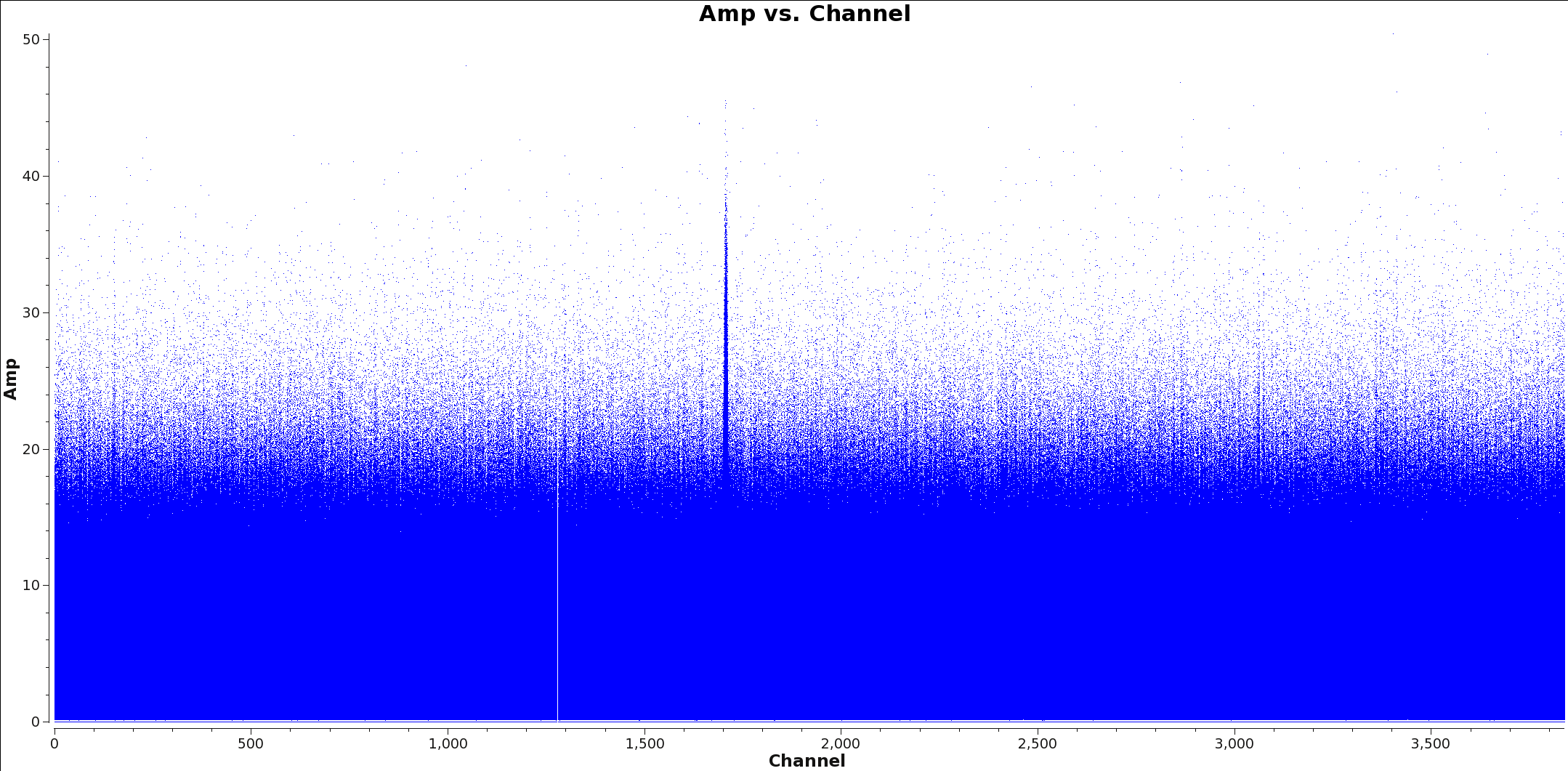
[[File:Calibrated_final_Field0_Spw0.png|thumb|Figure 1: spw 0]] | [[File:Calibrated_final_Field0_Spw0.png|thumb|Figure 1: spw 0]] | ||
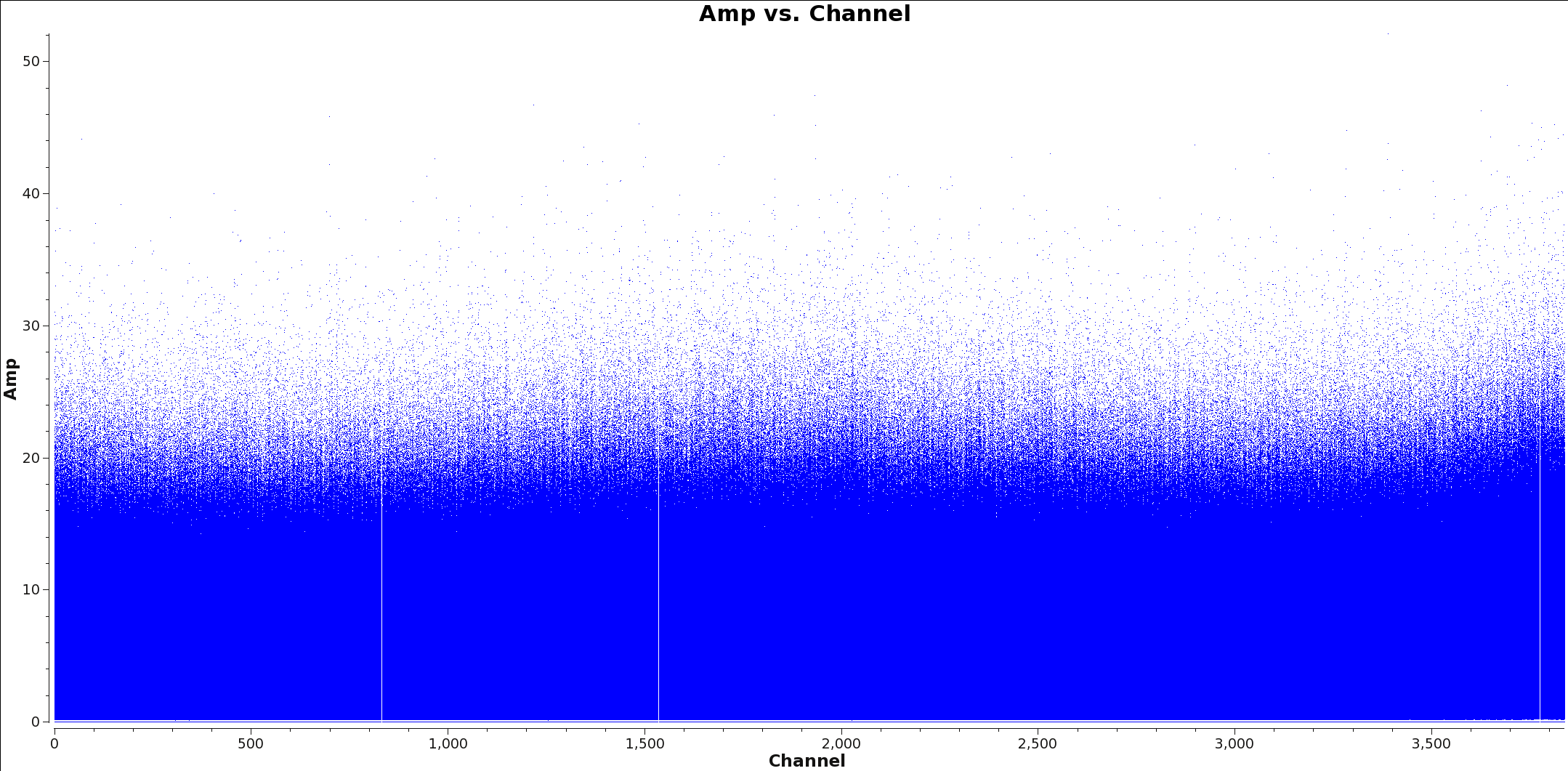
[[File:Calibrated_final_Field0_Spw3.png|thumb|Figure 2: Plots showing each spectral window for TW Hydra with channel vs amplitude. Spw 0 has a strong spectral line and spw 3 is a continuum only window.]] | [[File:Calibrated_final_Field0_Spw3.png|thumb|Figure 2: Plots showing each spectral window for TW Hydra with channel vs amplitude. Spw 0 has a strong spectral line and spw 3 is a continuum only window.]] | ||
Appropriate averaging of a measurement set can reduce overall data volume, making imaging faster with {{tclean}}. Since the continuum image is formed by essentially summing the entire bandwidth, we can spectrally average the input measurement set prior to imaging without significantly affecting the final imaging results. Below, we outline a procedure to create a spectrally averaged measurement set for continuum imaging. | |||
Appropriate averaging of a measurement set can reduce overall data volume, making imaging faster with {{ | |||
The first step is to identify which spectral windows (spws) you would like to include in the continuum. In general, you should include all spws with bandwidths greater than 250MHz to maximize the available continuum bandwidth. Once you have determined which spws you would like to use to form the continuum ms, set the contspw variable: | The first step is to identify which spectral windows (spws) you would like to include in the continuum. In general, you should include all spws with bandwidths greater than 250MHz to maximize the available continuum bandwidth. Once you have determined which spws you would like to use to form the continuum ms, set the contspw variable: | ||
| Line 28: | Line 26: | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
finalvis='calibrated_final.ms' # This is your output ms from the data | |||
# preparation script. | |||
# Set spws to be used to form continuum | # Set spws to be used to form continuum | ||
contspws = '0,1,2,3' | contspws = '0,1,2,3' | ||
| Line 36: | Line 36: | ||
To use the first method, create a channel vs. amplitude plot using plotms. An example command is given below. You may wish to change the averaging and the uvrange to identify extended emission. | To use the first method, create a channel vs. amplitude plot using plotms. An example command is given below. You may wish to change the averaging and the uvrange to identify extended emission. | ||
Figures 1 and 2 show example plots of | Figures 1 and 2 show example plots of a spw with and without lines to flag. Figure 1 shows a clear line at channel 1700 whereas Figure 2 shows no line emission and just continuum. | ||
Warning: If you apply channel averaging greater than 1, the numbers displayed on the channel axis will represent the channel bin number, rather than the averaged channel. | Warning: If you apply channel averaging greater than 1, the numbers displayed on the channel axis will represent the channel bin number, rather than the averaged channel. | ||
| Line 48: | Line 48: | ||
</source> | </source> | ||
The second, and more accurate, method is to use {{ | The second, and more accurate, method is to use {{tclean}} to make a quick dirty image cube of channels with niter set to zero and mode=’channel’. Inspecting this channel cube for line emission gives a better defined channel range to flag. The basic command to create dirty image cubes is given below. If you are going to use this method, you will need to set the [[Image_Continuum#Image_Parameters | imaging parameters]] before running the {{tclean}} command. This command should be repeated for each spw and science field. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 54: | Line 54: | ||
testimagename=’testImage’ | testimagename=’testImage’ | ||
field=[ | field=['0'] #list all target fields | ||
spw=[ | spw=['0','1','2','3'] #list all target spw’s | ||
for i in field: | for i in field: | ||
for j in spw: | for j in spw: | ||
tclean(vis=finalvis, | |||
imagename=testimagename+ | imagename=testimagename+'Field_'+str(i)+'_spw_'+str(j), | ||
field=str(i), | field=str(i), | ||
spw=str(j), | spw=str(j), | ||
# phasecenter=phasecenter, # uncomment if mosaic and set to appropriate field number | |||
# phasecenter='TRACKFIELD' # uncomment if imaging an ephemeris object, the phasecenter needs to be TRACKFIELD, not a field number as above. | |||
outframe= | specmode='cube', | ||
veltype=veltype, | |||
nchan=-1, | |||
outframe='REST', # velocity reference frame. See science goals | |||
niter=0, | niter=0, | ||
interactive=True, | interactive=True, | ||
| Line 72: | Line 75: | ||
weighting=weighting, | weighting=weighting, | ||
robust=robust, | robust=robust, | ||
pbcor=True, | |||
restoringbeam='common', | |||
gridder=gridder) | |||
</source> | </source> | ||
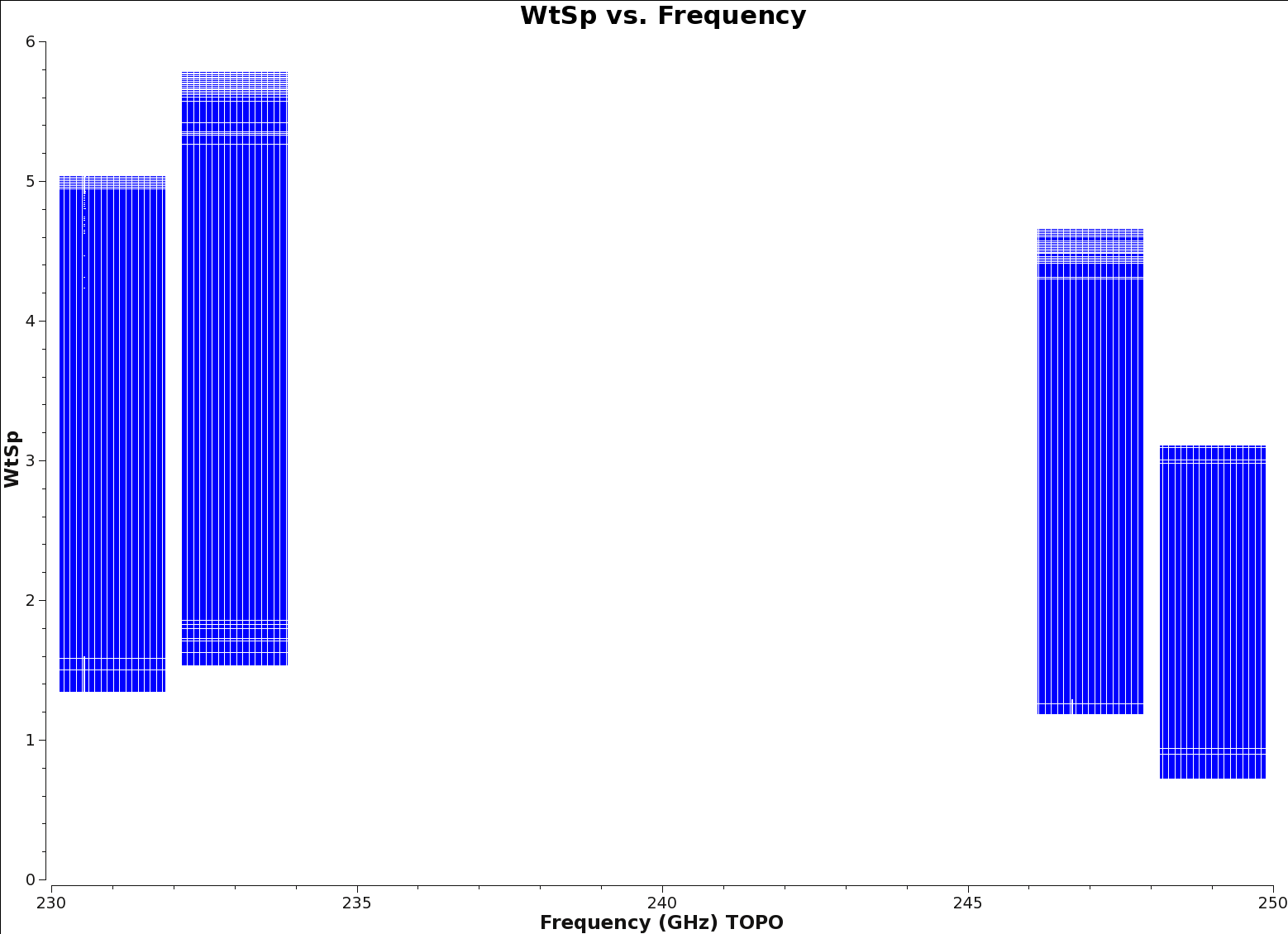
[[File:Wt_vs_Freq_spw0123.png|thumb|Figure 3: The weight spectrum vs Frequency for the calibrated_final.ms for each science spectral window. Notice how the weights in each individual spectral window do not vary wildly and have no outliers but the overall weight measure is different for each spectral window due to the different observing frequency.]] | [[File:Wt_vs_Freq_spw0123.png|thumb|Figure 3: The weight spectrum vs Frequency for the calibrated_final.ms for each science spectral window. Notice how the weights in each individual spectral window do not vary wildly and have no outliers but the overall weight measure is different for each spectral window due to the different observing frequency.]] | ||
The first step to flagging the spectral line channels in your data is to use the {{flagmanager}} task to save the state of the data before any flagging is applied. You will need to revert back to the non spectral line flagged dataset when line imaging is done later on. In addition, if you accidentally flag any data you can easily correct this by restoring the before_cont_flags file using the {{flagmanager}}. | The first step to flagging the spectral line channels in your data is to use the {{flagmanager}} task to save the state of the data before any flagging is applied. You will need to revert back to the non spectral line flagged dataset when line imaging is done later on. In addition, if you accidentally flag any data you can easily correct this by restoring the before_cont_flags file using the {{flagmanager}}. | ||
| Line 88: | Line 90: | ||
</source> | </source> | ||
Initialize the per-channel (or spectral) weights in the ms using [https://casa.nrao.edu/docs/TaskRef/initweights-task.html initweights]. This step ensures that when the flagged and unflagged channels are combined, the appropriate weighting is given to the final set of averaged channels. Figure 3 shows a representation of the weightspectrum vs. frequency plot you should see after initializing the spectral weights | Initialize the per-channel (or spectral) weights in the ms using [https://casa.nrao.edu/docs/TaskRef/initweights-task.html initweights]. This step ensures that when the flagged and unflagged channels are combined, the appropriate weighting is given to the final set of averaged channels. Figure 3 shows a representation of the weightspectrum vs. frequency plot you should see after initializing the spectral weights. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 102: | Line 104: | ||
</source> | </source> | ||
[[File:Calibrated_final_field0_spw0_LineChannelFlagged.png|thumb|Figure 4: Amp vs channel for spectral window 0 for TW Hydra with the spectral line flagged out.]] | [[File:Calibrated_final_field0_spw0_LineChannelFlagged.png|thumb|Figure 4: Amp vs channel for spectral window 0 for TW Hydra with the spectral line flagged out.]] | ||
Next, use the task {{flagdata}} to apply these flags. After this is done, check that the flags were applied correctly by using plotms to inspect the flagged ms. Figure 4 shows an example of what you should expect to see. | Next, use the task {{flagdata}} to apply these flags. After this is done, check that the flags were applied correctly by using plotms to inspect the flagged ms. Figure 4 shows an example of what you should expect to see. | ||
| Line 119: | Line 119: | ||
</source> | </source> | ||
Now | Now you can spectrally average the channels together to reduce the size of the continuum ms. The number of channels that you can average together is limited by what is referred to as "bandwidth smearing". When creating an interferometer image, we assume that the emission is essentially monochromatic. If you average large numbers of channels together, this is no longer an appropriate assumption and results in radial smearing in the image that increases in magnitude from the delay tracking center. For a detailed discussion, consult [http://adsabs.harvard.edu/abs/1999ASPC..180..371B Bridle and Schwab, 1999, Synthesis Imaging in Radio Astronomy II, 180, 371]. For a short derivation of the relevant quantities, see [https://safe.nrao.edu/wiki/pub/Main/RadioTutorial/BandwidthSmearing.pdf How to Calculate Bandwidth Smearing]. Conservative values for averaging channels in various ALMA bands are suggested below. As a rough guide, the number of channels you can average together is: | ||
* Bands 3, 4, 5, and 6: 125MHz/ (Channel Width (MHz)) | * Bands 3, 4, 5, and 6: 125MHz/ (Channel Width (MHz)) | ||
* | * Bands 7, 8, 9 and 10: 250MHz/ (Channel Width (MHz)) | ||
In general, you should make sure that the number of channels you are averaging together is an integer multiple of the original total number of channels. For example, if you have 128 channels and a channel width of 15.625 MHz in band 6, you can average together 8 channels at a time. The resulting ms will contain 16 final channels each with a channel width of 125MHz. | In general, you should make sure that the number of channels you are averaging together is an integer multiple of the original total number of channels. For example, if you have 128 channels and a channel width of 15.625 MHz in band 6, you can average together 8 channels at a time. The resulting ms will contain 16 final channels each with a channel width of 125MHz. | ||
[[Image:Bandwidthsmearingtable.png|center|frame|1200px]] ''This table lists the maximum bandwidth allowed for a reduction in peak response to a point source over the field of view of 1% for a a square and Gaussian bandpass for various observing frequencies and baselines for different two bandpass types.'' | If you want to be more careful, the following table will tell you the maximum bandwidth each averaged channel can be to avoid bandwidth smearing worse than about 1%. For example, at Band 7 (373 GHz) in a compact configuration (Bmax=500m) bandwidth smearing is relatively unimportant even for wide bandwidths (2.1 GHz), whereas for extended configurations (Bmax=10km) the maximum bandwidth at the same Band 7 should be of order 100 MHz or less. | ||
[[Image:Bandwidthsmearingtable.png|center|frame|1200px]] ''This table lists the maximum bandwidth allowed for a reduction in peak response to a point source over the field of view of 1% for a a square and Gaussian bandpass for various observing frequencies and baselines for different two bandpass types. See [https://safe.nrao.edu/wiki/pub/Main/RadioTutorial/BandwidthSmearing.pdf How to Calculate Bandwidth Smearing] for more information.'' | |||
Finally, the task {{split}} is used to average the channels together to produce the spectrally averaged continuum data set. | Finally, the task {{split}} is used to average the channels together to produce the spectrally averaged continuum data set. | ||
| Line 134: | Line 136: | ||
rmtables(contvis) | rmtables(contvis) | ||
os.system('rm -rf ' + contvis + '.flagversions') | os.system('rm -rf ' + contvis + '.flagversions') | ||
split(vis=finalvis, | |||
spw=contspws, | |||
outputvis=contvis, | |||
width=[256,8,8,8], # number of channels to average together. The final channel width should be less than 125MHz in Bands 3, 4, and 6 | |||
# and 250MHz in Bands 7, 8, 9 and 10. | |||
datacolumn='data') | |||
</source> | </source> | ||
Now you should check the weights of the new continuum measurement set. The ratio of the weights value between Time Domain Mode (TDM) and Frequency Domain Mode (FDM) windows should be approximately equal to the ratio of the channel widths. In addition, any heavily flagged channels should have their weights scaled by the ratio of the unflagged bandwidth to the bandwidth per output channel. For more information about data weights, see the [https://casaguides.nrao.edu/index.php/DataWeightsAndCombination Data Weights and Combination] guide. | Now you should check the weights of the new continuum measurement set. The ratio of the weights value between Time Domain Mode (TDM) and Frequency Domain Mode (FDM) windows should be approximately equal to the ratio of the channel widths. For more information on the correlator modes TDM and FDM, see section 5.1 of the [https://almascience.nrao.edu/documents-and-tools/cycle5/alma-technical-handbook ALMA Technical Handbook]. In addition, any heavily flagged channels should have their weights scaled by the ratio of the unflagged bandwidth to the bandwidth per output channel. For more information about data weights, see the [https://casaguides.nrao.edu/index.php/DataWeightsAndCombination Data Weights and Combination] guide. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 150: | Line 153: | ||
Finally, we need to use the {{flagmanager}} tasks to restore the ms file to its original unflagged state, so that later we can do continuum subtraction and line imaging. | Finally, we need to use the {{flagmanager}} tasks to restore the ms file to its original unflagged state, so that later we can do continuum subtraction and line imaging. | ||
[[File:Amp_vs_uvdist.png|thumb|Figure 5: Amplitude vs UVDistance plot of the continuum, colored by spw.]] | |||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
| Line 172: | Line 173: | ||
Before imaging, you should set a few key parameters that you will use throughout the rest of the script. | Before imaging, you should set a few key parameters that you will use throughout the rest of the script. | ||
Set the field id for the science target you are interested in imaging. You can use the listobs file generated during the imaging prep step to find this information. If you are imaging a mosaic then you will have to select all the fields. For example, field = ‘3~25’. You can also set the field variable to the name of the source, as long as it matches the name in the listobs file. Do not leave the field parameter blank. If you leave the field parameter blank, | Set the field id for the science target you are interested in imaging. You can use the listobs file generated during the imaging prep step to find this information. If you are imaging a mosaic then you will have to select all the fields. For example, field = ‘3~25’. You can also set the field variable to the name of the source, as long as it matches the name in the listobs file. Do not leave the field parameter blank. If you leave the field parameter blank, {{tclean}} will attempt to image all sources in the measurement set. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 180: | Line 181: | ||
</source> | </source> | ||
Uncomment the | Uncomment the gridder that is relevant to your dataset. Set the phase center by field id or coordinates if you are imaging a mosaic. Check the spatial setup in the weblog (for pipeline calibrations) or qa2 report (for manual calibrations) to find the phase center. You should choose the central field for the phase center in order to get the best results. | ||
If you are unsure which field to use for the phase center after looking at the weblog then you may use the following Analysis Utilities command: | If you are unsure which field to use for the phase center after looking at the weblog then you may use the following Analysis Utilities command: | ||
<source lang="python"> | <source lang="python"> | ||
au.pickCellSize( | au.pickCellSize('calibrated_final.ms',imsize=True). | ||
</source> | </source> | ||
This will return a four element array with that contains the calculated cell size, the X axis number of pixels, the Y axis number of pixels, and the field number that is most centered in the mosaic. You may use this as the phase center field id in your script. If you haven't installed Analysis Utilities, see [https://casaguides.nrao.edu/index.php?title=Analysis_Utilities Obtaining Analysis Utilities] for instructions. | This will return a four element array with that contains the calculated cell size, the X axis number of pixels, the Y axis number of pixels, and the field number that is most centered in the mosaic. You may use this as the phase center field id in your script. If you haven't installed Analysis Utilities, see [https://casaguides.nrao.edu/index.php?title=Analysis_Utilities Obtaining Analysis Utilities] for instructions. | ||
Another method of finding the central field number uses the [https://casaguides.nrao.edu/index.php/Plotmosaic plotmosaic] | Another method of finding the central field number uses the Analysis Utilities [https://casaguides.nrao.edu/index.php/Plotmosaic plotmosaic] au.plotmosaic(‘calibrated_final.ms’). This method will produce a plot of all fields with their corresponding field numbers plotted on the sky. You can also set the phase center with the coordinates at the center of this plot. | ||
For ephemeris objects such as planets, the outframe should be set to phasecenter = 'TRACKFIELD', not a field number as above. | |||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
# | # gridder='standard' # uncomment if single field | ||
# | # gridder='mosaic' # uncomment if mosaic or if combining one 7m and one 12m pointing. | ||
# phasecenter=3 # uncomment and set to field number for phase | # phasecenter=3 # uncomment and set to field number for phase | ||
# center. Note lack of ''. Use the weblog to | |||
# determine which pointing to use. Remember that the | |||
# field ids for each pointing will be re-numbered | |||
# after your initial split. You can also specify the | |||
# phase center using coordinates, e.g., | |||
# phasecenter='J2000 19h30m00 -40d00m00'. | |||
# phasecenter = 'TRACKFIELD' # If imaging an ephemeris object (planet, etc), the phasecenter needs to be TRACKFIELD, not a field number as above. | |||
</source> | </source> | ||
[[File:Calibrated_final_AmpVsUVWave.png|thumb|Figure 6: Amplitude versus UV wave for the calibrated_final.ms showing the maximum uv wave point for determining cell size.]] | [[File:Calibrated_final_AmpVsUVWave.png|thumb|Figure 6: Amplitude versus UV wave for the calibrated_final.ms showing the maximum uv wave point for determining cell size.]] | ||
Next, determine the cell (or pixel) size. To do this, you need to determine the approximate resolution of your observations. Then you divide the resolution in arcsec by 5 to 8 to adequately sample the PSF. The resolution of a particular interferometer observations can be estimated from the length of the longest baseline: | Next, determine the cell (or pixel) size. To do this, you need to determine the approximate resolution of your observations. Then you divide the resolution in arcsec by 5 to 8 to adequately sample the PSF. The resolution of a particular interferometer observations can be estimated from the length of the longest baseline: | ||
| Line 226: | Line 229: | ||
<pre style="background-color: #E0FFFF;"> | <pre style="background-color: #E0FFFF;"> | ||
cellsize(arc seconds) = resolution(arcsec)/ | cellsize(arc seconds) = resolution(arcsec)/7 | ||
</pre> | </pre> | ||
| Line 233: | Line 236: | ||
If this is a single field, the image size can usually be approximated to be the same size as the primary beam of the telescope. The ALMA 12m primary beam in arcsec scales as 6300 / nu[GHz] and the ALMA 7m primary beam in arcsec scales as 10608 / nu[GHz], where nu[GHz] is the sky frequency. However, if there is significant point source and/or extended emission beyond the edges of your initial images, you should increase the imsize to incorporate more emission. | If this is a single field, the image size can usually be approximated to be the same size as the primary beam of the telescope. The ALMA 12m primary beam in arcsec scales as 6300 / nu[GHz] and the ALMA 7m primary beam in arcsec scales as 10608 / nu[GHz], where nu[GHz] is the sky frequency. However, if there is significant point source and/or extended emission beyond the edges of your initial images, you should increase the imsize to incorporate more emission. | ||
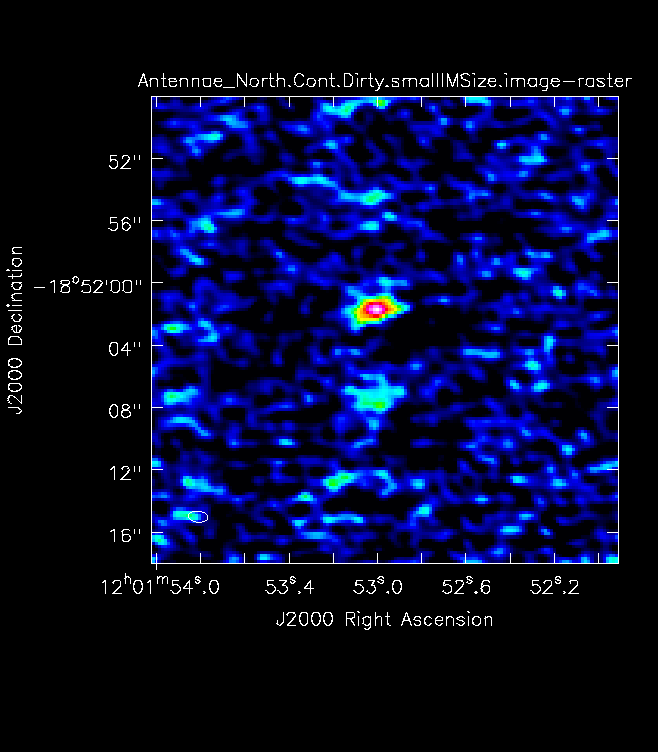
For mosaics, make the image substantially larger than the mosaic footprint. Use au.plotmosaic(finalvis) to find the mosaic footprint in arcseconds. Padding the imsize substantially avoids artifacts in the image. You should be able to see the edges of the outside fields being cut off in an appropriately padded image. | For mosaics, make the image substantially larger than the mosaic footprint. Use au.plotmosaic(finalvis) to find the mosaic footprint in arcseconds. Padding the imsize substantially avoids artifacts in the image. You should be able to see the edges of the outside fields being cut off in an appropriately padded image. In Figure 7 you will see a mosaic field under padded with the edges not visible. Figure 8 shows an image made with a larger imsize of the same mosaic with the edges clearly visible. | ||
[[File:Antennae_Antennae_North.Cont.Dirty.image.png|thumb|Figure 7]] | [[File:Antennae_Antennae_North.Cont.Dirty.image.png|thumb|Figure 7]] | ||
[[File:Antennae_North.Cont.Dirty.smallIMSize.image.png|thumb|Figure 8: A mosaic of Antennae showing under padding and correctly padded images. For the correctly padded image you can see the edges of the primary beam field whereas the under padded does not cut off.]] | [[File:Antennae_North.Cont.Dirty.smallIMSize.image.png|thumb|Figure 8: A mosaic of Antennae showing under padding and correctly padded images. For the correctly padded image you can see the edges of the primary beam field whereas the under padded does not cut off.]] | ||
Note that the imsize parameter is in PIXELS, not arcsec, so you will need to divide the image size in arcsec by the pixel size to determine a value for imsize. | Note that the imsize parameter is in PIXELS, not arcsec, so you will need to divide the image size in arcsec by the pixel size to determine a value for imsize. | ||
| Line 250: | Line 251: | ||
</source> | </source> | ||
When imaging you will need to set two specific velocity parameters called outframe and veltype. Outframe is the coordinate system used for the observation. If you have access to the original proposal, this can be found in the Observing Tool (OT) under field setup. A list of acceptable outframes that can be used in CASA can be found at https://help.almascience.org/ | When imaging lines, you will need to set two specific velocity parameters called outframe and veltype. Outframe is the coordinate system used for the observation. If you have access to the original proposal, this can be found in the Observing Tool (OT) under field setup. A list of acceptable outframes that can be used in CASA can be found at https://help.almascience.org/kb/articles/what-are-the-frequency-reference-frames-in-casa. Note: heliocentric(hel) is deprecated in CASA. Use barycentric(bary) in this case. The most common choices are 'bary' and 'lsrk'. Usually 'bary' is used for sources where z>0.2 ('extragalactic") and 'lsrk is used for 'galactic' sources. For ephemeris objects, the outframe should be set to a blank string, for example outframe = <nowiki>''</nowiki>, as you have likely already regridded to the source velocity in cvel() or can allow tclean to do it on the fly. | ||
You will also have to set the veltype for the {{ | You will also have to set the veltype for the {{tclean}} command. This variable has only two options available, radio and optical. Due to an interaction between the ALMA Observing Tool and CASA, set the veltype to radio. Even if the object has an optically defined velocity, the sensitivity calculation uses the radio definition. This will avoid confusion in comparing the achieved sensitivity to the expected sensitivity. | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
outframe=' | outframe='lsrk' # velocity reference frame. See science goals. | ||
veltype='radio' # velocity type. | veltype='radio' # velocity type. | ||
</source> | </source> | ||
The last four parameters that must be set are associated with how {{ | The last four parameters that must be set are associated with how {{tclean}} will weight and clean the data. During the imaging process, the individual visibilities are placed on a uv grid and then combined. Weighting determines how {{tclean}} will combine the uv gridded to produce an image. There are several weighting schemes that can be used: | ||
* Natural: Each cell in the uv plane is weighted by the number of points in the cell. This weighting scheme is the default in {{ | * Natural: Each cell in the uv plane is weighted by the number of points in the cell. This weighting scheme is the default in {{tclean}}. It results in a lower noise image at the expense of worse angular resolution. | ||
* Uniform: Each cell in the uv plane has the same weight. In this case, the sidelobes will be reduced because the imaging plane is filled in more uniformly. This weighting scheme also gives more weight to longer baselines resulting in higher resolution at the expense of higher noise. It can emphasize visibilities with calibration errors. | * Uniform: Each cell in the uv plane has the same weight. In this case, the sidelobes will be reduced because the imaging plane is filled in more uniformly. This weighting scheme also gives more weight to longer baselines resulting in higher resolution at the expense of higher noise. It can emphasize visibilities with calibration errors. | ||
* Briggs: This weighting scheme is a combination of natural and uniform weighting. The weighting is controlled by the robust parameter. A robust parameter of 2 gives natural weighting, -2 uniform weighting, and a number in between a combination of natural and uniform. Refer to [ | * Briggs: This weighting scheme is a combination of natural and uniform weighting. The weighting is controlled by the robust parameter. A robust parameter of 2 gives natural weighting, -2 uniform weighting, and a number in between a combination of natural and uniform. Refer to [https://casadocs.readthedocs.io/en/stable/notebooks/memo-series.html?highlight=briggs#Dan-Briggs'-Dissertation---Robust-Weighting Brigg's Thesis] for more information. Currently, robust is set to 0.5, which gives a good compromise between natural and uniform weighting. You may find, after imaging, that you have to decrease the noise or angular resolution based on the science goals. Playing with the robust parameter can affect your final noise in the image and also the angular resolution. If you are making a mosaic image do not use a robust value smaller than 0 as this may introduce major artifacts in the images including uneven noise across the image. If you choose to do any form of uvtapering to the data in tclean(), set robust to 2 (Natural weighting) to avoid upweighting points that are going to be downweighted by uv-taper. | ||
The parameters niter and threshold provide two ways to stop the {{ | The parameters niter and threshold provide two ways to stop the {{tclean}} process. The niter parameter is the maximum number of iterations allowed. After this limit has been reached, {{tclean}} will terminate. The threshold parameter sets a flux density threshold for the {{tclean}}. When the peak residual is below this value, {{tclean}} terminates. When cleaning interactively, we recommend setting the niter parameter to a large(say 1000), but not too large value (say 10000 or 100000) so that {{tclean}} terminates eventually in the case of human error. The threshold can be left at the default of 0.0mJy for interactive tclean. For non-interactive tclean, the recommendation is to set the threshold to several times the noise in the final image. To determine the threshold for cube images, run tclean with niter set to zero and inspect the resulting image in the {{viewer}}. In a line-free channel, select a region and look at the statistics panel to determine the noise level. For more detailed instructions, see the “Image the Spectral Line Data” section of [https://casaguides.nrao.edu/index.php?title=EVLA_Spectral_Line_Imaging_Analysis_IRC%2B10216 EVLA Spectral Line Imaging Analysis IRC+10216]. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 278: | Line 279: | ||
</source> | </source> | ||
For more information on the various options available in {{ | For more information on the various options available in {{tclean}}, refer to [https://science.nrao.edu/science/meetings/2016/15th-synthesis-imaging-workshop/documents/wilner_vla16.pdf David Wilner’s Imaging and Deconvolution presentation] from the 2016 Synthesis Imaging Workshop. The table below provides helpful pointers to relevant slides. | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 294: | Line 295: | ||
== Imaging the Continuum == | == Imaging the Continuum == | ||
[[File:Cont_before_clean.png|thumb|Figure 9: During interactive {{tclean}}, the GUI will show an initial dirty image. From this GUI, create the {{tclean}} mask.]] | |||
[[File:Cont_before_clean.png|thumb|Figure 9: During interactive {{ | |||
[[File:Cont_before_clean_mask.png|thumb|Figure 10: After a mask is created, the green arrow will be illuminated. Press this to begin the first 100 iterations. Once cleaning is complete, press the red "X" to finish.]] | [[File:Cont_before_clean_mask.png|thumb|Figure 10: After a mask is created, the green arrow will be illuminated. Press this to begin the first 100 iterations. Once cleaning is complete, press the red "X" to finish.]] | ||
[[File:Final_Cont_residual.png|thumb|Figure 11: The final residual image.]] | [[File:Final_Cont_residual.png|thumb|Figure 11: The final residual image.]] | ||
Now that you have set all of the imaging parameters you will need in {{tclean}}, you can proceed to imaging the continuum. The [https://casaguides.nrao.edu/index.php/First_Look_at_Imaging First Look at Imaging CASAGuide] gives an introduction to cleaning and imaging. You may also find the [https://casaguides.nrao.edu/index.php/Automasking_Guide Automasking Guide] useful in allowing {{tclean}} to generate the mask used for cleaning automatically. | |||
We provide an abbreviated set of commands here. | |||
If you are imaging a mosaic, the phasecenter parameter should be set. Refer to the [[Image_Continuum#Image Parameters | Image Parameters]] section of this guide for instructions on how to determine this for your project. Type “help | If you are imaging a mosaic, the phasecenter parameter should be set. Refer to the [[Image_Continuum#Image Parameters | Image Parameters]] section of this guide for instructions on how to determine this for your project. Type “help tclean()” in CASA if you would like to explore the possible parameters of {{tclean}}. Specode=’mfs’ sets the spectral gridding type to multi-frequency synthesis and creates a continuum image. If you are imaging the aggregate continuum in Band 3 or 4 and have a fractional bandwidth larger than 10%, you should consider using multi-term multi-frequency synthesis (deconvolver='mtmfs' and nterms=2). This tclean mode accounts for spatial spectral index variations and especially important to include if you have a spatially resolved, high S/N source. For more information on multi-term multi-frequency synthesis, see [https://www.aanda.org/articles/aa/pdf/2011/08/aa17104-11.pdf Rau, U., & Cornwell, T.J. 2011, A&A, 532, AA71]. | ||
Clean interactively as the threshold is set at 0 mJy. The mask parameter may also be added if you have an existing file. You can create a mask from the dirty image using the instructions at [[Create_a_Clean_Mask_from_Continuum_Image_or_Moment_Cube]]. | |||
In CASA 5.4 and later, {{tclean}} calls with gridder = 'mosaic' have an additional parameter mosweight with a default of True. When mosweight = True, the gridder weights each field in the mosaic independently. The mosweight parameter is particularly important for mosaics with non-uniform sensitivity, with rectangular shapes, or when using more uniform values of robust Briggs weighting. For more information on mosweight, please see the {{tclean}} documentation. | |||
<source lang="python"> | <source lang="python"> | ||
| Line 316: | Line 314: | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
tclean(vis=contvis, | |||
imagename=contimagename, | |||
field=field, | |||
# | # phasecenter=phasecenter, # uncomment if mosaic or imaging an ephemeris object | ||
# mosweight=True, # uncomment if mosaic | |||
specmode='mfs', | |||
deconvolver='hogbom', | |||
# Uncomment the below to image with nterms>1. Use if fractional bandwidth is >10%. | |||
#deconvolver='mtmfs', | |||
#nterms=2, | |||
imsize = imsize, | |||
cell= cell, | |||
weighting = weighting, | |||
robust = robust, | |||
niter = niter, | |||
threshold = threshold, | |||
interactive = True, | |||
gridder = gridder, | |||
pbcor = True, | |||
usepointing=False) | |||
</source> | </source> | ||
Figure 9 shows the {{ | Figure 9 shows the {{tclean}} GUI that will appear. If no source is apparent, no cleaning should be done. Press the red “X” to complete the task. If a source is apparent, create a mask around it using the mouse. Once a mask is created, the green arrow will be illuminated and you can begin the first round of cleaning. Figure 10 shows what a mask would look like for our example source. The logger window gives you vital information on the progression of {{tclean}}. Once the cycle is complete, the residuals will appear in the GUI. You should continue to iterate until the region inside the mask matches the noise outside the mask. You may need multiple cycles depending on the complexity of the source. Figure 11 shows an example of when we chose to stop cleaning. | ||
The red "X" will stop {{ | The red "X" will stop {{tclean}} where you are, the blue arrow will stop the interactive part of {{tclean}}, but continue to clean non-interactively until reaching the number of iterations requested (niter) or the flux density threshold (whichever comes first), and the green circle arrow will clean until it reaches the "iterations" parameter on the left side of the green area. These are the only safe exit buttons to use during {{tclean}}. DO NOT CTRL-C OR KILL TCLEAN WHILE IT IS RUNNING. If you do this, it is very likely that your ms will be corrupted. | ||
[[Image:Final_Continuum_Image.png|center|frame|800px]] ''The final continuum image.'' | [[Image:Final_Continuum_Image.png|center|frame|800px]] ''The final continuum image.'' | ||
| Line 340: | Line 344: | ||
After creating the continuum image, check the noise and resolution to make sure the results match the expected values. Use the [https://almascience.nrao.edu/proposing/sensitivity-calculator ALMA Sensitivity Calculator] to estimate the expected sensitivity. | After creating the continuum image, check the noise and resolution to make sure the results match the expected values. Use the [https://almascience.nrao.edu/proposing/sensitivity-calculator ALMA Sensitivity Calculator] to estimate the expected sensitivity. | ||
For each image it creates, {{ | For each image it creates, {{tclean}} generates several images with the name imagename+extension. If you re-run tclean with the same imagename, {{tclean}} will use the existing files as a starting point, continuing the tclean where you left off. To start completely from scratch, either change the imagename or delete all the files from the previous {{tclean}} run. Note that CASA retains some image information in memory, so to truly delete the images, you need to run the {{rmtables}} command. You will also want to remove datacolumns that were added by tclean with {{clearcal}} and {{delmod}}. See below for an example. | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
for ext in [ | clearcal(contvis) | ||
delmod(contvis) | |||
for ext in ['.image','.mask','.model','.image.pbcor','.psf','.residual','.pb','.sumwt']: | |||
rmtables(contimagename+ext) | |||
</source> | </source> | ||
Once you are happy with your continuum image(s), you can continue to '''[[Self_Calibration_Template | Self-Calibration Template]]''' or '''[[Image_Line | Spectral Line Imaging Template]]'''. If you do not wish to self-calibrate or create line cubes, continue with this guide to create primary beam corrected images and fits files. | Once you are happy with your continuum image(s), you can continue to '''[[Self_Calibration_Template | Self-Calibration Template]]''' or '''[[Image_Line | Spectral Line Imaging Template]]'''. If you do not wish to self-calibrate or create line cubes, continue with this guide to create primary beam corrected images and fits files. | ||
== Export the images == | == Export the images == | ||
| Line 382: | Line 368: | ||
exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True) | exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True) | ||
myimages = glob.glob("*. | myimages = glob.glob("*.pb") | ||
for image in myimages: | for image in myimages: | ||
exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True) | exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True) | ||
</source> | </source> | ||
| Line 395: | Line 382: | ||
# in CASA | # in CASA | ||
os.system("rm -rf *.png") | os.system("rm -rf *.png") | ||
mycontimages = glob.glob(" | mycontimages = glob.glob("*mfs*manual.image") | ||
for cimage in mycontimages: | for cimage in mycontimages: | ||
mymax=imstat(cimage)['max'][0] | |||
mymin=-0.1*mymax | |||
outimage = cimage+'.png' | outimage = cimage+'.png' | ||
os.system('rm -rf '+outimage) | os.system('rm -rf '+outimage) | ||
imview(raster={'file':cimage,'range':[ | imview(raster={'file':cimage,'range':[mymin,mymax]},out=outimage) | ||
</source> | |||
'''[[Guide_NA_ImagingTemplate | Return to the Main Page]]''' | '''[[Guide_NA_ImagingTemplate | Return to the Main Page]]''' | ||
Latest revision as of 20:33, 28 February 2024
This guide should be used after completing Prepare the data for Imaging. You should have created calibrated_final.ms prior to proceeding. Commands for this guide can be found in scriptForImaging_template.py available on github.
Check CASA version
This template is for use in CASA versions 6.1.1.15 and greater. The following code checks the version of CASA and exits if it is less than 6.1.1.15. You can download the appropriate version of CASA from Obtaining CASA . See Prepare for Imaging section on the main page of this guide for more information.
# in CASA
import re
try:
import casalith
except:
print("Script requires CASA 6.0 or greater")
if casalith.compare_version("<",[6,1,1,15]):
print("Please use CASA version greater than or equal to 6.1.1-15 with this script")
Create an Averaged Continuum MS
Appropriate averaging of a measurement set can reduce overall data volume, making imaging faster with tclean. Since the continuum image is formed by essentially summing the entire bandwidth, we can spectrally average the input measurement set prior to imaging without significantly affecting the final imaging results. Below, we outline a procedure to create a spectrally averaged measurement set for continuum imaging.
The first step is to identify which spectral windows (spws) you would like to include in the continuum. In general, you should include all spws with bandwidths greater than 250MHz to maximize the available continuum bandwidth. Once you have determined which spws you would like to use to form the continuum ms, set the contspw variable:
# in CASA
finalvis='calibrated_final.ms' # This is your output ms from the data
# preparation script.
# Set spws to be used to form continuum
contspws = '0,1,2,3'
The next step is to identify and flag the spectral lines in all spectral windows that you will use to make the continuum image. You will split and average the data with the spectral lines flagged so that your final spectrally averaged continuum ms only contains continuum emission. Two methods commonly used to identical spectral line emission are: 1) a channel vs. amplitude plot of the visibilities and 2) a dirty image of the cube.
To use the first method, create a channel vs. amplitude plot using plotms. An example command is given below. You may wish to change the averaging and the uvrange to identify extended emission.
Figures 1 and 2 show example plots of a spw with and without lines to flag. Figure 1 shows a clear line at channel 1700 whereas Figure 2 shows no line emission and just continuum.
Warning: If you apply channel averaging greater than 1, the numbers displayed on the channel axis will represent the channel bin number, rather than the averaged channel.
# in CASA
plotms(vis=finalvis, xaxis='channel', yaxis='amplitude',
ydatacolumn='data',
avgtime='1e8', avgscan=True, avgchannel='1',
iteraxis='spw' )
The second, and more accurate, method is to use tclean to make a quick dirty image cube of channels with niter set to zero and mode=’channel’. Inspecting this channel cube for line emission gives a better defined channel range to flag. The basic command to create dirty image cubes is given below. If you are going to use this method, you will need to set the imaging parameters before running the tclean command. This command should be repeated for each spw and science field.
#In CASA
testimagename=’testImage’
field=['0'] #list all target fields
spw=['0','1','2','3'] #list all target spw’s
for i in field:
for j in spw:
tclean(vis=finalvis,
imagename=testimagename+'Field_'+str(i)+'_spw_'+str(j),
field=str(i),
spw=str(j),
# phasecenter=phasecenter, # uncomment if mosaic and set to appropriate field number
# phasecenter='TRACKFIELD' # uncomment if imaging an ephemeris object, the phasecenter needs to be TRACKFIELD, not a field number as above.
specmode='cube',
veltype=veltype,
nchan=-1,
outframe='REST', # velocity reference frame. See science goals
niter=0,
interactive=True,
cell=cell,
imsize=imsize,
weighting=weighting,
robust=robust,
pbcor=True,
restoringbeam='common',
gridder=gridder)
The first step to flagging the spectral line channels in your data is to use the flagmanager task to save the state of the data before any flagging is applied. You will need to revert back to the non spectral line flagged dataset when line imaging is done later on. In addition, if you accidentally flag any data you can easily correct this by restoring the before_cont_flags file using the flagmanager.
# in CASA
flagmanager(vis=finalvis,mode='save',
versionname='before_cont_flags')
Initialize the per-channel (or spectral) weights in the ms using initweights. This step ensures that when the flagged and unflagged channels are combined, the appropriate weighting is given to the final set of averaged channels. Figure 3 shows a representation of the weightspectrum vs. frequency plot you should see after initializing the spectral weights.
# in CASA
initweights(vis=finalvis,wtmode='weight',dowtsp=True)
The exact spectral window and channel ranges to flag in the flagchannels variable needs to be specified. For example, if spws 2&3 have a line between channels 1201 and 2199 and spectral windows 0 and 1 are line-free the command would be:
# in CASA
flagchannels='2:1201~2199,3:1201~2199' # modify the channel range for your dataset
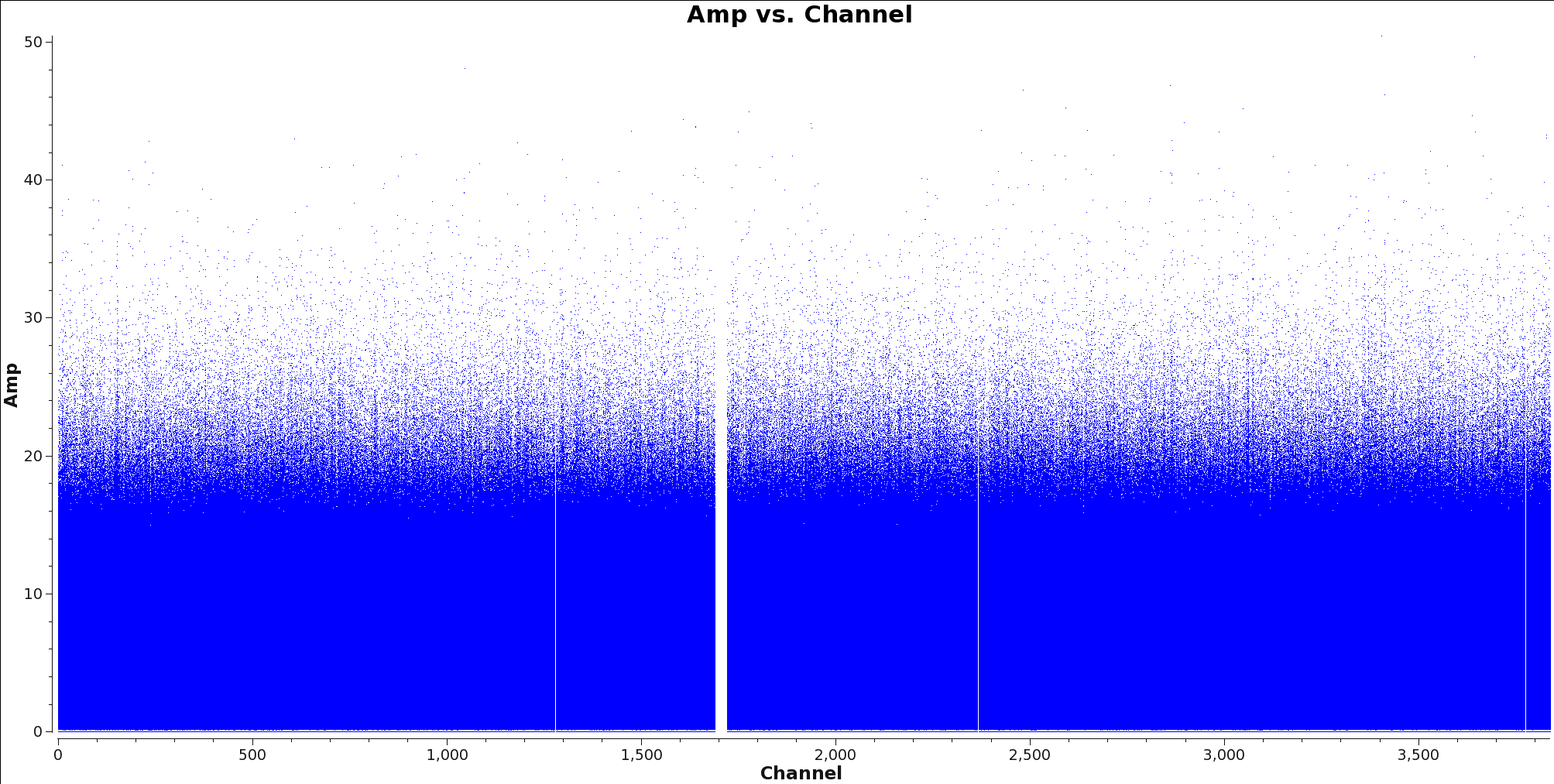
Next, use the task flagdata to apply these flags. After this is done, check that the flags were applied correctly by using plotms to inspect the flagged ms. Figure 4 shows an example of what you should expect to see.
# in CASA
flagdata(vis=finalvis,mode='manual',
spw=flagchannels,flagbackup=False)
# check that flags are as expected, NOTE must check reload on plotms
# gui if its still open.
plotms(vis=finalvis,yaxis='amp',xaxis='channel',
avgchannel='1',avgtime='1e8',avgscan=True,iteraxis='spw')
Now you can spectrally average the channels together to reduce the size of the continuum ms. The number of channels that you can average together is limited by what is referred to as "bandwidth smearing". When creating an interferometer image, we assume that the emission is essentially monochromatic. If you average large numbers of channels together, this is no longer an appropriate assumption and results in radial smearing in the image that increases in magnitude from the delay tracking center. For a detailed discussion, consult Bridle and Schwab, 1999, Synthesis Imaging in Radio Astronomy II, 180, 371. For a short derivation of the relevant quantities, see How to Calculate Bandwidth Smearing. Conservative values for averaging channels in various ALMA bands are suggested below. As a rough guide, the number of channels you can average together is:
- Bands 3, 4, 5, and 6: 125MHz/ (Channel Width (MHz))
- Bands 7, 8, 9 and 10: 250MHz/ (Channel Width (MHz))
In general, you should make sure that the number of channels you are averaging together is an integer multiple of the original total number of channels. For example, if you have 128 channels and a channel width of 15.625 MHz in band 6, you can average together 8 channels at a time. The resulting ms will contain 16 final channels each with a channel width of 125MHz.
If you want to be more careful, the following table will tell you the maximum bandwidth each averaged channel can be to avoid bandwidth smearing worse than about 1%. For example, at Band 7 (373 GHz) in a compact configuration (Bmax=500m) bandwidth smearing is relatively unimportant even for wide bandwidths (2.1 GHz), whereas for extended configurations (Bmax=10km) the maximum bandwidth at the same Band 7 should be of order 100 MHz or less.
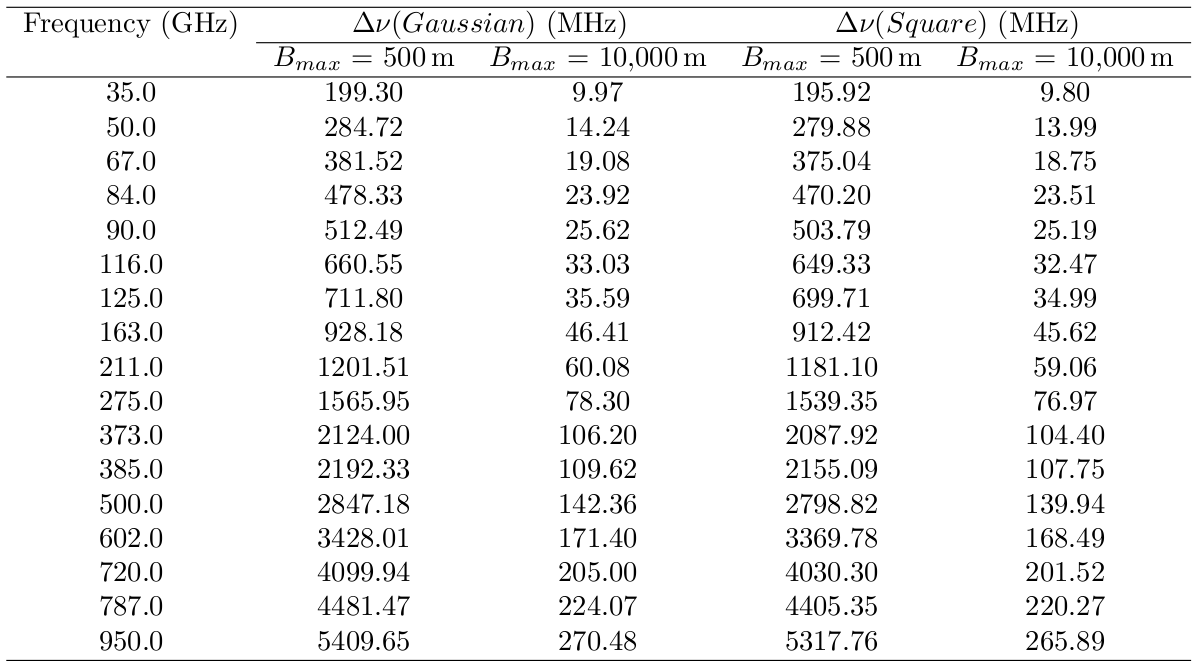
This table lists the maximum bandwidth allowed for a reduction in peak response to a point source over the field of view of 1% for a a square and Gaussian bandpass for various observing frequencies and baselines for different two bandpass types. See How to Calculate Bandwidth Smearing for more information.
Finally, the task split is used to average the channels together to produce the spectrally averaged continuum data set.
# in CASA
contvis='calibrated_final_cont.ms'
rmtables(contvis)
os.system('rm -rf ' + contvis + '.flagversions')
split(vis=finalvis,
spw=contspws,
outputvis=contvis,
width=[256,8,8,8], # number of channels to average together. The final channel width should be less than 125MHz in Bands 3, 4, and 6
# and 250MHz in Bands 7, 8, 9 and 10.
datacolumn='data')
Now you should check the weights of the new continuum measurement set. The ratio of the weights value between Time Domain Mode (TDM) and Frequency Domain Mode (FDM) windows should be approximately equal to the ratio of the channel widths. For more information on the correlator modes TDM and FDM, see section 5.1 of the ALMA Technical Handbook. In addition, any heavily flagged channels should have their weights scaled by the ratio of the unflagged bandwidth to the bandwidth per output channel. For more information about data weights, see the Data Weights and Combination guide.
# in CASA
# update the antenna and field parameters for your dataset
plotms(vis=contvis, yaxis='wtsp',xaxis='freq',spw='',antenna='DA42',field='0')
Finally, we need to use the flagmanager tasks to restore the ms file to its original unflagged state, so that later we can do continuum subtraction and line imaging.
# in CASA
# If you flagged any line channels, restore the previous flags
flagmanager(vis=finalvis,mode='restore',
versionname='before_cont_flags')
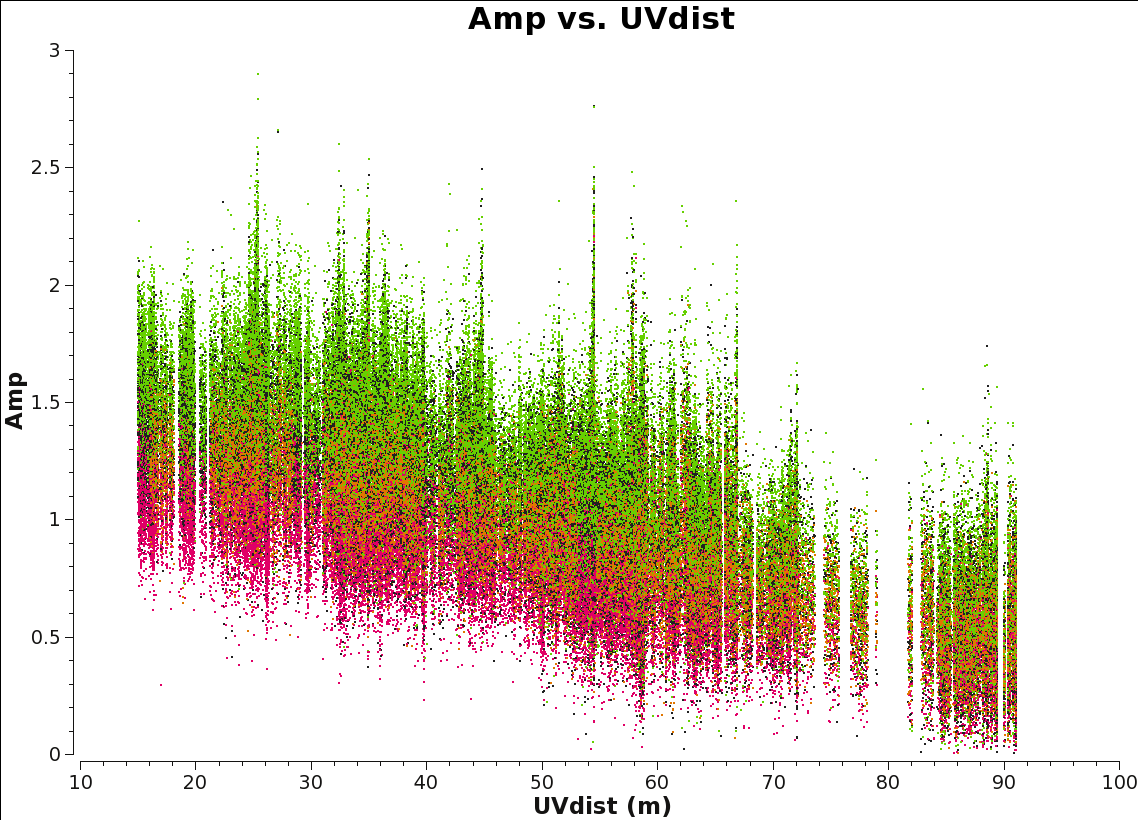
It is a good practice to inspect the final continuum ms to make sure that it is correct. In general, the distribution of amplitude vs. uv distance should be smooth. Recall this plot will be a horizontal line for a point source, while the short uv-distances will have higher amplitudes for a more extended object.
# in CASA
plotms(vis=contvis,xaxis='uvdist',yaxis='amp',coloraxis='spw')
Image Parameters
Before imaging, you should set a few key parameters that you will use throughout the rest of the script.
Set the field id for the science target you are interested in imaging. You can use the listobs file generated during the imaging prep step to find this information. If you are imaging a mosaic then you will have to select all the fields. For example, field = ‘3~25’. You can also set the field variable to the name of the source, as long as it matches the name in the listobs file. Do not leave the field parameter blank. If you leave the field parameter blank, tclean will attempt to image all sources in the measurement set.
# in CASA
# update for your ms
field='0'
Uncomment the gridder that is relevant to your dataset. Set the phase center by field id or coordinates if you are imaging a mosaic. Check the spatial setup in the weblog (for pipeline calibrations) or qa2 report (for manual calibrations) to find the phase center. You should choose the central field for the phase center in order to get the best results.
If you are unsure which field to use for the phase center after looking at the weblog then you may use the following Analysis Utilities command:
au.pickCellSize('calibrated_final.ms',imsize=True).
This will return a four element array with that contains the calculated cell size, the X axis number of pixels, the Y axis number of pixels, and the field number that is most centered in the mosaic. You may use this as the phase center field id in your script. If you haven't installed Analysis Utilities, see Obtaining Analysis Utilities for instructions.
Another method of finding the central field number uses the Analysis Utilities plotmosaic au.plotmosaic(‘calibrated_final.ms’). This method will produce a plot of all fields with their corresponding field numbers plotted on the sky. You can also set the phase center with the coordinates at the center of this plot.
For ephemeris objects such as planets, the outframe should be set to phasecenter = 'TRACKFIELD', not a field number as above.
# in CASA
# gridder='standard' # uncomment if single field
# gridder='mosaic' # uncomment if mosaic or if combining one 7m and one 12m pointing.
# phasecenter=3 # uncomment and set to field number for phase
# center. Note lack of ''. Use the weblog to
# determine which pointing to use. Remember that the
# field ids for each pointing will be re-numbered
# after your initial split. You can also specify the
# phase center using coordinates, e.g.,
# phasecenter='J2000 19h30m00 -40d00m00'.
# phasecenter = 'TRACKFIELD' # If imaging an ephemeris object (planet, etc), the phasecenter needs to be TRACKFIELD, not a field number as above.
Next, determine the cell (or pixel) size. To do this, you need to determine the approximate resolution of your observations. Then you divide the resolution in arcsec by 5 to 8 to adequately sample the PSF. The resolution of a particular interferometer observations can be estimated from the length of the longest baseline:
resolution (radian) ~ (observed wavelength) / (length of longest baseline)
If the baseline is measured in wavelengths, this becomes
resolution (radian) ~ 1.0 / (length of longest baseline in wavelengths)
To convert from radians to arcsec, we multiple by 206265.0 to obtain:
resolution(arcsec) ~ 206265.0/(longest baseline in wavelengths)
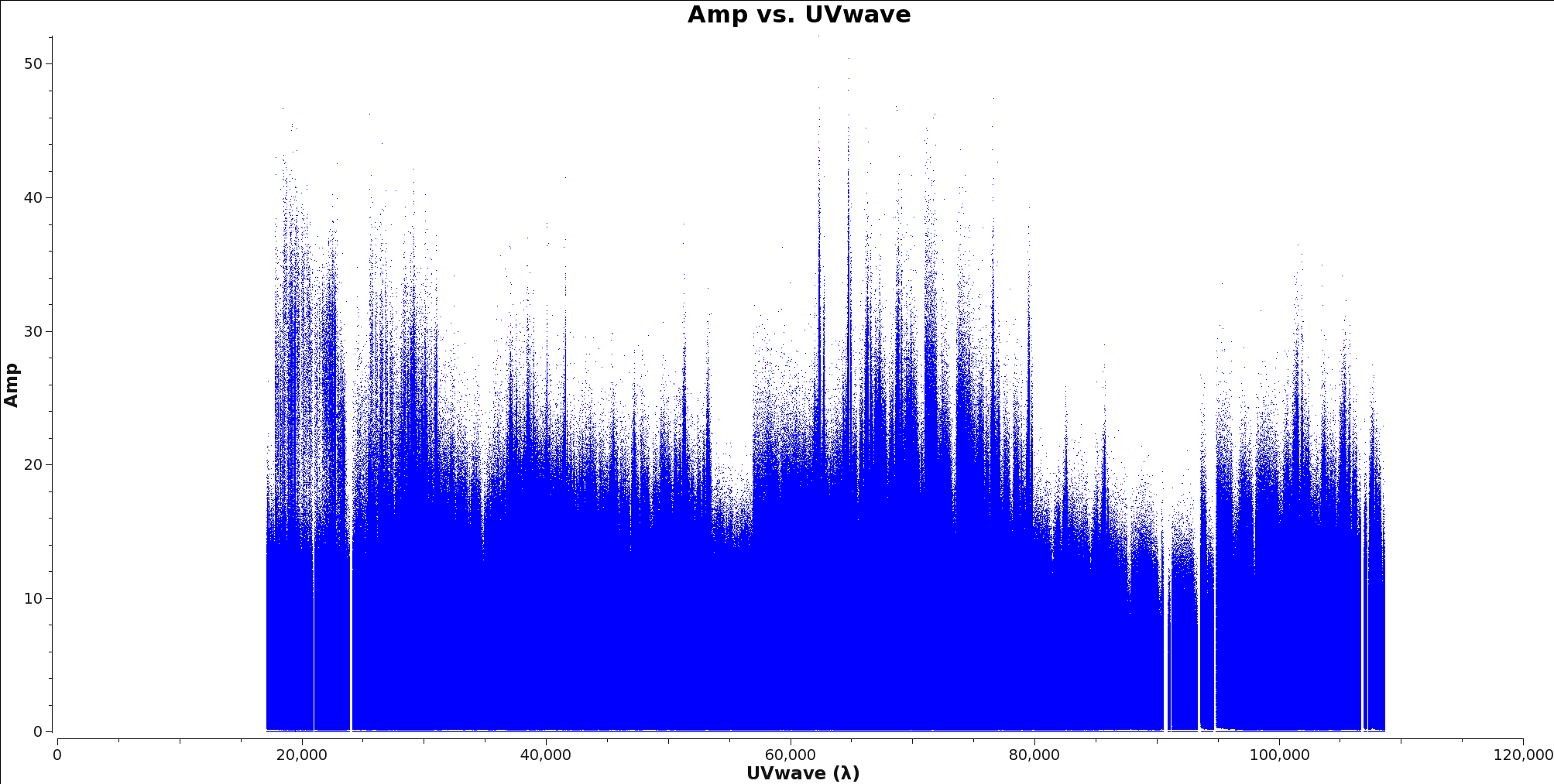
To determine the longest baseline in wavelengths use plotms with xaxis=’uvwave’ and yaxis=’amp’ on your ms file. Figure 6 shows an example of this plot. It is generally better to oversample your beam than to undersample, particularly for observations with poor uv-coverage.
cellsize(arc seconds) = resolution(arcsec)/7
The next step is to determine the image size in pixels. There are two methods of doing this depending on if you are imaging one field or a mosaic.
If this is a single field, the image size can usually be approximated to be the same size as the primary beam of the telescope. The ALMA 12m primary beam in arcsec scales as 6300 / nu[GHz] and the ALMA 7m primary beam in arcsec scales as 10608 / nu[GHz], where nu[GHz] is the sky frequency. However, if there is significant point source and/or extended emission beyond the edges of your initial images, you should increase the imsize to incorporate more emission.
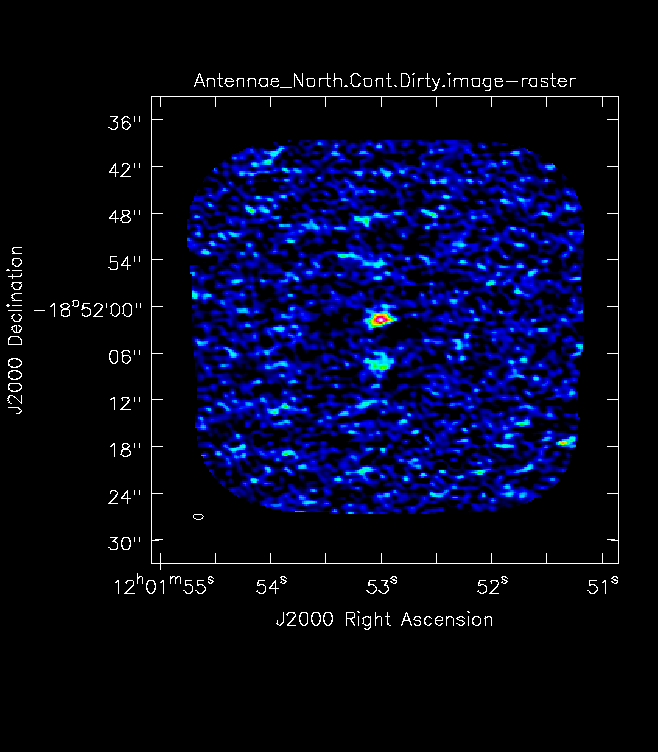
For mosaics, make the image substantially larger than the mosaic footprint. Use au.plotmosaic(finalvis) to find the mosaic footprint in arcseconds. Padding the imsize substantially avoids artifacts in the image. You should be able to see the edges of the outside fields being cut off in an appropriately padded image. In Figure 7 you will see a mosaic field under padded with the edges not visible. Figure 8 shows an image made with a larger imsize of the same mosaic with the edges clearly visible.
Note that the imsize parameter is in PIXELS, not arcsec, so you will need to divide the image size in arcsec by the pixel size to determine a value for imsize.
If you would like to check any of these calculations you may use the following command, au.pickCellSize('calibrated_final.ms', imsize=True), in CASA. If you haven't installed Analysis Utilities, see Obtaining Analysis Utilities for instructions. This will return a four element array of the calculated cell size, the x axis imsize, the y axis imsize, and the central field id number.
# in CASA
cell='1arcsec' # cell size for imaging.
imsize = [128,128] # size of image in pixels.
When imaging lines, you will need to set two specific velocity parameters called outframe and veltype. Outframe is the coordinate system used for the observation. If you have access to the original proposal, this can be found in the Observing Tool (OT) under field setup. A list of acceptable outframes that can be used in CASA can be found at https://help.almascience.org/kb/articles/what-are-the-frequency-reference-frames-in-casa. Note: heliocentric(hel) is deprecated in CASA. Use barycentric(bary) in this case. The most common choices are 'bary' and 'lsrk'. Usually 'bary' is used for sources where z>0.2 ('extragalactic") and 'lsrk is used for 'galactic' sources. For ephemeris objects, the outframe should be set to a blank string, for example outframe = '', as you have likely already regridded to the source velocity in cvel() or can allow tclean to do it on the fly.
You will also have to set the veltype for the tclean command. This variable has only two options available, radio and optical. Due to an interaction between the ALMA Observing Tool and CASA, set the veltype to radio. Even if the object has an optically defined velocity, the sensitivity calculation uses the radio definition. This will avoid confusion in comparing the achieved sensitivity to the expected sensitivity.
# in CASA
outframe='lsrk' # velocity reference frame. See science goals.
veltype='radio' # velocity type.
The last four parameters that must be set are associated with how tclean will weight and clean the data. During the imaging process, the individual visibilities are placed on a uv grid and then combined. Weighting determines how tclean will combine the uv gridded to produce an image. There are several weighting schemes that can be used:
- Natural: Each cell in the uv plane is weighted by the number of points in the cell. This weighting scheme is the default in tclean. It results in a lower noise image at the expense of worse angular resolution.
- Uniform: Each cell in the uv plane has the same weight. In this case, the sidelobes will be reduced because the imaging plane is filled in more uniformly. This weighting scheme also gives more weight to longer baselines resulting in higher resolution at the expense of higher noise. It can emphasize visibilities with calibration errors.
- Briggs: This weighting scheme is a combination of natural and uniform weighting. The weighting is controlled by the robust parameter. A robust parameter of 2 gives natural weighting, -2 uniform weighting, and a number in between a combination of natural and uniform. Refer to Brigg's Thesis for more information. Currently, robust is set to 0.5, which gives a good compromise between natural and uniform weighting. You may find, after imaging, that you have to decrease the noise or angular resolution based on the science goals. Playing with the robust parameter can affect your final noise in the image and also the angular resolution. If you are making a mosaic image do not use a robust value smaller than 0 as this may introduce major artifacts in the images including uneven noise across the image. If you choose to do any form of uvtapering to the data in tclean(), set robust to 2 (Natural weighting) to avoid upweighting points that are going to be downweighted by uv-taper.
The parameters niter and threshold provide two ways to stop the tclean process. The niter parameter is the maximum number of iterations allowed. After this limit has been reached, tclean will terminate. The threshold parameter sets a flux density threshold for the tclean. When the peak residual is below this value, tclean terminates. When cleaning interactively, we recommend setting the niter parameter to a large(say 1000), but not too large value (say 10000 or 100000) so that tclean terminates eventually in the case of human error. The threshold can be left at the default of 0.0mJy for interactive tclean. For non-interactive tclean, the recommendation is to set the threshold to several times the noise in the final image. To determine the threshold for cube images, run tclean with niter set to zero and inspect the resulting image in the viewer. In a line-free channel, select a region and look at the statistics panel to determine the noise level. For more detailed instructions, see the “Image the Spectral Line Data” section of EVLA Spectral Line Imaging Analysis IRC+10216.
# in CASA
weighting = 'briggs'
robust=0.5
niter=1000
threshold = '0.0mJy'
For more information on the various options available in tclean, refer to David Wilner’s Imaging and Deconvolution presentation from the 2016 Synthesis Imaging Workshop. The table below provides helpful pointers to relevant slides.
| Pixel and Image Size | Slides 40-41 |
| Weighting | Slides 42-48, 60-61 |
| Deconvolution Algorithms | Slides 50-53, 62 |
Imaging the Continuum
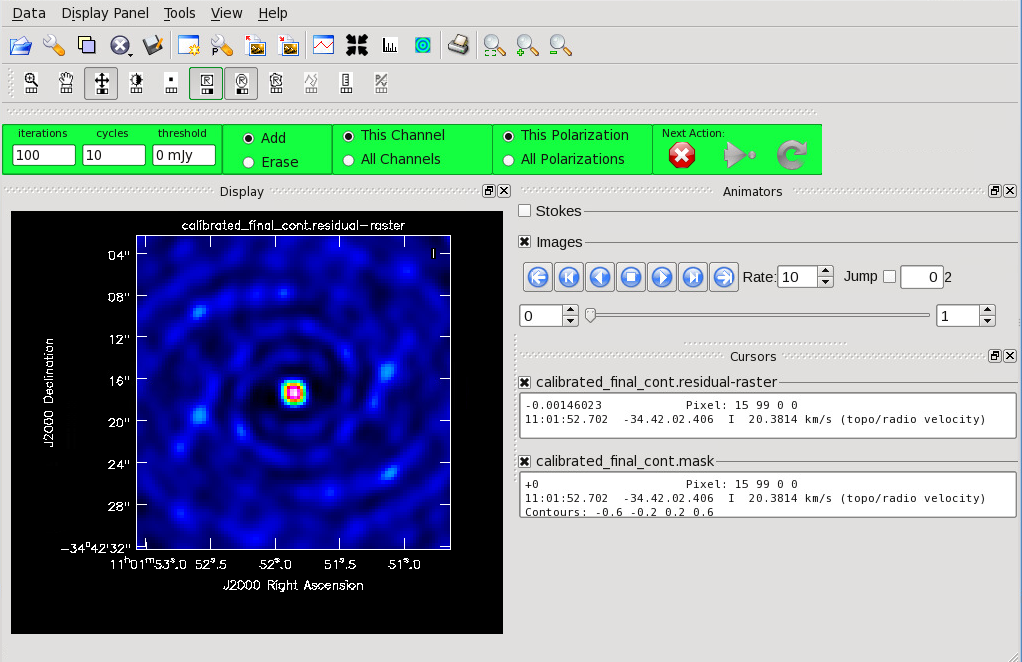
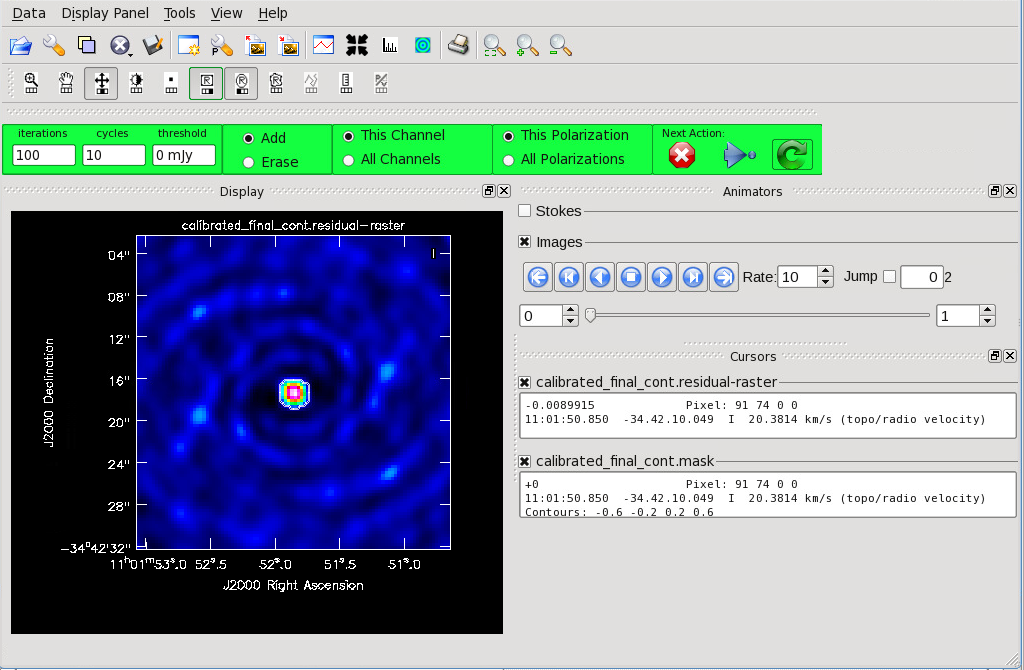
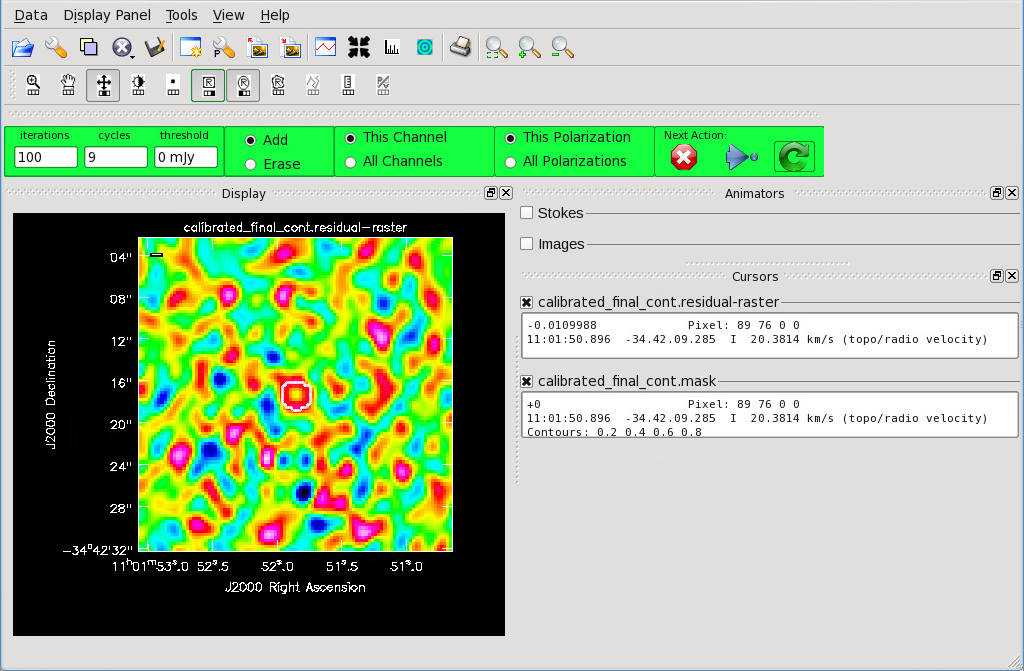
Now that you have set all of the imaging parameters you will need in tclean, you can proceed to imaging the continuum. The First Look at Imaging CASAGuide gives an introduction to cleaning and imaging. You may also find the Automasking Guide useful in allowing tclean to generate the mask used for cleaning automatically.
We provide an abbreviated set of commands here. If you are imaging a mosaic, the phasecenter parameter should be set. Refer to the Image Parameters section of this guide for instructions on how to determine this for your project. Type “help tclean()” in CASA if you would like to explore the possible parameters of tclean. Specode=’mfs’ sets the spectral gridding type to multi-frequency synthesis and creates a continuum image. If you are imaging the aggregate continuum in Band 3 or 4 and have a fractional bandwidth larger than 10%, you should consider using multi-term multi-frequency synthesis (deconvolver='mtmfs' and nterms=2). This tclean mode accounts for spatial spectral index variations and especially important to include if you have a spatially resolved, high S/N source. For more information on multi-term multi-frequency synthesis, see Rau, U., & Cornwell, T.J. 2011, A&A, 532, AA71. Clean interactively as the threshold is set at 0 mJy. The mask parameter may also be added if you have an existing file. You can create a mask from the dirty image using the instructions at Create_a_Clean_Mask_from_Continuum_Image_or_Moment_Cube.
In CASA 5.4 and later, tclean calls with gridder = 'mosaic' have an additional parameter mosweight with a default of True. When mosweight = True, the gridder weights each field in the mosaic independently. The mosweight parameter is particularly important for mosaics with non-uniform sensitivity, with rectangular shapes, or when using more uniform values of robust Briggs weighting. For more information on mosweight, please see the tclean documentation.
# in CASA
contvis = 'calibrated_final_cont.ms'
contimagename = 'calibrated_final_cont'
# in CASA
tclean(vis=contvis,
imagename=contimagename,
field=field,
# phasecenter=phasecenter, # uncomment if mosaic or imaging an ephemeris object
# mosweight=True, # uncomment if mosaic
specmode='mfs',
deconvolver='hogbom',
# Uncomment the below to image with nterms>1. Use if fractional bandwidth is >10%.
#deconvolver='mtmfs',
#nterms=2,
imsize = imsize,
cell= cell,
weighting = weighting,
robust = robust,
niter = niter,
threshold = threshold,
interactive = True,
gridder = gridder,
pbcor = True,
usepointing=False)
Figure 9 shows the tclean GUI that will appear. If no source is apparent, no cleaning should be done. Press the red “X” to complete the task. If a source is apparent, create a mask around it using the mouse. Once a mask is created, the green arrow will be illuminated and you can begin the first round of cleaning. Figure 10 shows what a mask would look like for our example source. The logger window gives you vital information on the progression of tclean. Once the cycle is complete, the residuals will appear in the GUI. You should continue to iterate until the region inside the mask matches the noise outside the mask. You may need multiple cycles depending on the complexity of the source. Figure 11 shows an example of when we chose to stop cleaning.
The red "X" will stop tclean where you are, the blue arrow will stop the interactive part of tclean, but continue to clean non-interactively until reaching the number of iterations requested (niter) or the flux density threshold (whichever comes first), and the green circle arrow will clean until it reaches the "iterations" parameter on the left side of the green area. These are the only safe exit buttons to use during tclean. DO NOT CTRL-C OR KILL TCLEAN WHILE IT IS RUNNING. If you do this, it is very likely that your ms will be corrupted.
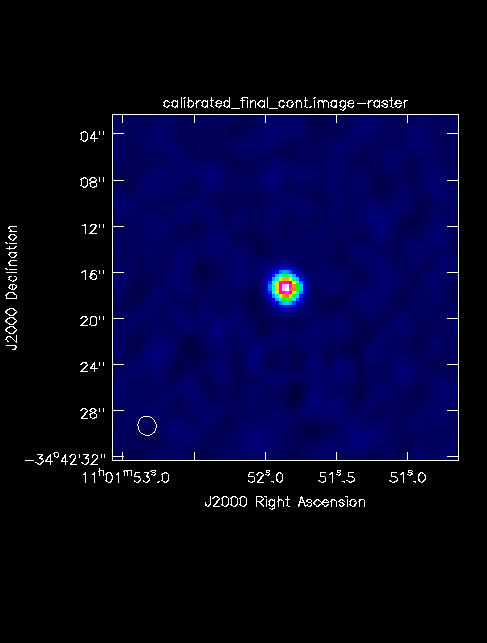
The final continuum image.
After creating the continuum image, check the noise and resolution to make sure the results match the expected values. Use the ALMA Sensitivity Calculator to estimate the expected sensitivity.
For each image it creates, tclean generates several images with the name imagename+extension. If you re-run tclean with the same imagename, tclean will use the existing files as a starting point, continuing the tclean where you left off. To start completely from scratch, either change the imagename or delete all the files from the previous tclean run. Note that CASA retains some image information in memory, so to truly delete the images, you need to run the rmtables command. You will also want to remove datacolumns that were added by tclean with clearcal and delmod. See below for an example.
# in CASA
clearcal(contvis)
delmod(contvis)
for ext in ['.image','.mask','.model','.image.pbcor','.psf','.residual','.pb','.sumwt']:
rmtables(contimagename+ext)
Once you are happy with your continuum image(s), you can continue to Self-Calibration Template or Spectral Line Imaging Template. If you do not wish to self-calibrate or create line cubes, continue with this guide to create primary beam corrected images and fits files.
Export the images
Use exportfits to create fits files of the *.flux and *.pbcor files.
# in CASA
import glob
myimages = glob.glob("*.pbcor")
for image in myimages:
exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True)
myimages = glob.glob("*.pb")
for image in myimages:
exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True)
Create Diagnostic PNGs
The First Look at Image Analysis guide gives an introduction to a variety of options to begin image analysis, including using immoments to create moment maps. The following commands create png files of the continuum image.
# in CASA
os.system("rm -rf *.png")
mycontimages = glob.glob("*mfs*manual.image")
for cimage in mycontimages:
mymax=imstat(cimage)['max'][0]
mymin=-0.1*mymax
outimage = cimage+'.png'
os.system('rm -rf '+outimage)
imview(raster={'file':cimage,'range':[mymin,mymax]},out=outimage)