M100 Band3 ACA 4.2.2: Difference between revisions
| (218 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
<div style="background-color:#E0FFFF;border:4px solid #FF9966;"> | |||
<div style="font-size:250%; color:red; text-align:center;"> | |||
This page is currently under construction. | |||
</div> | |||
<div style="font-size:200%; color:black; text-align:center"> | |||
DO NOT USE IT. | |||
</div> | |||
<div style="font-size:150%; color:black; text-align:center"> | |||
To navigate the CASAguides pages, visit [http://casaguides.nrao.edu/ | |||
casaguides.nrao.edu | |||
] | |||
</div> | |||
</div> | |||
[[Category:ALMA]][[Category:Calibration]][[Category:Spectral Line]] | [[Category:ALMA]][[Category:Calibration]][[Category:Spectral Line]] | ||
*'''This guide requires CASA 4.1.0 and assumes that you have downloaded | *'''This guide requires CASA 4.1.0 and assumes that you have downloaded M100_Band3_7m_UncalibratedData.tgz from [[M100_Band3#Obtaining_the_Data]]''' | ||
*'''Details of the ALMA observations are provided at [[M100_Band3]] | *'''Details of the ALMA observations are provided at [[M100_Band3]] | ||
| Line 11: | Line 24: | ||
==Overview== | ==Overview== | ||
This part of the M100 Band 3 7m CASA guide will | This part of the M100 Band 3 7m CASA guide will take you through the process of fully calibrating the visibility data. We will begin by flagging (marking as bad) data known to be useless before any inspection, for example data where one telescope blocks the line of sight of another. Then we will apply telescope-generated calibration tables to partially correct for atmospheric effects. After inspecting the data, we will flag some additional data that exhibit pathologies. Then we will use observations of the calibrators Titan, 3c273, and J1215+1654 to derive the phase and amplitude response of individual antennas as a function of time and frequency ("phase", "amplitude", and "bandpass" calibrations). We will apply these to the data and then extract the calibrated source data into a file appropriate for imaging. | ||
The general procedure in this guide follows the other ALMA CASA guides: [[NGC3256Band3]] and [[TWHydraBand7]]. | The general procedure in this guide follows the other ALMA CASA guides: [[IRAS16293Band9]], [[NGC3256Band3]] and [[TWHydraBand7]]. | ||
==Unpack the Data== | ==Unpack the Data== | ||
Once you have downloaded the | Once you have downloaded the M100_Band3_7m_UncalibratedData.tgz, unpack the file in a terminal outside CASA using | ||
<source lang="bash"> | <source lang="bash"> | ||
tar -xvzf | tar -xvzf M100_Band3_7m_UncalibratedData.tgz | ||
</source> | </source> | ||
| Line 29: | Line 42: | ||
<source lang="bash"> | <source lang="bash"> | ||
cd | cd M100_Band3_7m_UncalibratedData | ||
</source> | </source> | ||
| Line 41: | Line 54: | ||
</source> | </source> | ||
to look at the files present. | to look at the files present. Note that a description of the observations can be found here: [[M100_Band3]] | ||
The files are in native ALMA format (ALMA Science Data Model or ASDM) and will be converted to measurement sets using the CASA task {{importasdm}} below. | |||
This guide requires Python module [[analysis_Utilities|analysisUtils]]. If you have not already installed [[analysis_Utilities|analysisUtils]] please follow the link to do so. | This guide requires Python module [[analysis_Utilities|analysisUtils]]. If you have not already installed [[analysis_Utilities|analysisUtils]] please follow the link to do so. | ||
| Line 53: | Line 68: | ||
</source> | </source> | ||
Be sure that you are using the version indicated at the top of this page. | Be sure that you are using the correct CASA version, indicated at the top of this page. | ||
==Confirm your version of CASA== | ==Confirm your version of CASA== | ||
| Line 77: | Line 92: | ||
for a full description and download instructions. Analysis Utilities are updated frequently so if its been a while since you installed it, its probably worth doing it again. If you are at an ALMA site or ARC, the analysis utilities are probably already installed and up to date. | for a full description and download instructions. Analysis Utilities are updated frequently so if its been a while since you installed it, its probably worth doing it again. If you are at an ALMA site or ARC, the analysis utilities are probably already installed and up to date. | ||
== A Note on Setting up to use multiple datasets == | |||
We will calibrate the six datasets individually and concatenate them into a single dataset at the end, before proceeding with the imaging part. | |||
To do this, we start by defining and array called "basename" consisting of the uid names of the six data files. This will allow us to step through the calibration of the six datasets, where appropriate, using these 'for' loops. We will not be able to show every diagnostic plot but we give an example of each and the syntax to generate the rest. Also, please be aware that even on a very fast machine this whole process can take a while, as we are dealing with a lot of data. | |||
You only need to define your list of files once per CASA session. Then "basename" will be a variable in the casapy shell. You can check if it exists by typing "print basename". In the interests of allowing you to easily exit and restart CASA and pick this guide up at any point we will redefine "basename" in each section of the guide. Feel free to skip this step if you've already defined it in your session. Remember that if you log out of CASA and want to continue with the calibration using these arrays, you will have to re-issue this command. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
# Define | # Define an array containing the 'uid' names of all of our data sets | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
# | for name in basename: | ||
print name | |||
</source> | |||
== Create Measurement Sets == | |||
The raw data have been provided to you in the ASDM format. It is the native format of the data produced by the observatory. | |||
Before we can proceed to the calibration, we will need to convert those data to the CASA MS format. This is done simply with the task importasdm. | |||
<source lang="python"> | |||
#In CASA | |||
for name in basename: | |||
importasdm(asdm = name, asis='*') | |||
</source> | |||
==Initial Inspection== | |||
We will take stock of what we have and obtain summary information about each dataset (targets observed, time range, spectral setup, and so on) by using the task {{listobs}} to output .txt format files. If you have not already done so, begin by reviewing the description of the observations here: [[M100_Band3]]. | |||
<source lang="python"> | |||
#In CASA | |||
# Loop over each element in the list and create summary file using listobs | # Loop over each element in the list and create summary file using listobs | ||
for | for name in basename: | ||
os.system('rm '+ | os.system('rm -rf '+name+'.ms.listobs.txt') | ||
listobs(vis= | listobs(vis=name+'.ms', listfile=name+'.ms.listobs.txt', verbose=True) | ||
</source> | </source> | ||
The "for" loop executes for each element in basename, calling listobs and directing the output to a file called, e.g., "uid___A002_X5e971a_X124.ms.listobs.txt" for the first measurement set. | |||
Note that after cutting and pasting a 'for' loop like this you often have to press return twice to execute. You may also want to take care to paste a line at a time if you are having trouble copy and pasting. Even better, you can use "cpaste" to paste blocks of code. To do so type "cpaste" at the CASA prompt, paste your commands, and then type "--" and hit return on the final (otherwise empty) line. This should look something like this: | Note that after cutting and pasting a 'for' loop like this you often have to press return twice to execute. You may also want to take care to paste a line at a time if you are having trouble copy and pasting. Even better, you can use "cpaste" to paste blocks of code. To do so type "cpaste" at the CASA prompt, paste your commands, and then type "--" and hit return on the final (otherwise empty) line. This should look something like this: | ||
| Line 116: | Line 146: | ||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
: | : | ||
:for | :for name in basename: | ||
: print | : print name | ||
:-- | :-- | ||
uid___A002_X5e971a_X124 | uid___A002_X5e971a_X124 | ||
| Line 132: | Line 162: | ||
cpaste should be ''much'' more robust than copying-and-pasting directly into the shell but if you have trouble, just carefully paste one line at a time directly into CASA and hit return until the desired command executes. | cpaste should be ''much'' more robust than copying-and-pasting directly into the shell but if you have trouble, just carefully paste one line at a time directly into CASA and hit return until the desired command executes. | ||
You can browse through the listobs output as you would normally look at a text file (use emacs, vi, or another editor). You can also send the output to the terminal from inside of CASA. To do so type: | |||
<source lang="python"> | <source lang="python"> | ||
| Line 450: | Line 480: | ||
</pre> | </pre> | ||
This output shows that | This output shows that four sources were observed in each data set: J1229+0203(3c273), Titan, J1215+1654, and M100. | ||
* | * '''M100''' is the science target. Note that the source corresponds to a number of individual fields (see the Field ID column). There are 23 pointings for the M100 Mosaic. | ||
* '''Titan''' is observed once and | * '''Titan''' is observed once and is supposed to be used to set the absolute flux scale of the data (missing in dataset X4eb). However, fluxes are not determined due to bad quality, we will not use Titan as an amplitude calibrator. | ||
* '''J1229+0203(3c273)''' plays two roles: it will serve as our bandpass calibrator, to characterize the frequency response of the antennas, and will be used to set the absolute flux scale of the data. | * '''J1229+0203(3c273)''' plays two roles: it will serve as our bandpass calibrator, to characterize the frequency response of the antennas, and will be used to set the absolute flux scale of the data. | ||
* '''J1215+1654''' is "gain calibrator" to track changes in the phase and amplitude response of the telescopes over time. Observations of J1215+1654 are interleaved with observations of the M100. | * '''J1215+1654''' is "gain calibrator" to track changes in the phase and amplitude response of the telescopes over time. Observations of J1215+1654 are interleaved with observations of the M100 mosaic. | ||
The output also shows that the data contain many spectral windows. Using the labeling scheme in the listobs above these are: | The output also shows that the data contain many spectral windows. Using the labeling scheme in the listobs above these are: | ||
* '''spw 16''','''spw 18''','''spw 20''', and '''spw 22''' hold our science data. These are "Frequency Domain Mode" (FDM) data with small (0.49 MHz) channel width and wide total bandwidth. As a result these have a lot of channels (4080). spw 22 holds the upper sideband (USB) data and includes the CO(1-0) line. We will focus on these data. (datasets X3f3 and X5b3 only have two science spectral windows: '''spw 8''' and '''spw 10''') | |||
* '''spw 16''','''spw 18''','''spw 20''', and '''spw 22''' hold our science data. These are "Frequency Domain Mode" (FDM) data with small (0.49 MHz) channel width and wide | * '''spw 8''', '''spw 10''', '''spw 12''', and '''spw 14''' hold lower a resolution processing ("Time Domain Mode", TDM) of the data from the same part of the spectrum (baseband). These data have only 128 channels across 2 GHz bandwidth and so have a much coarser channel spacing than the FDM data. These are used to create the Tsys calibration tables. (datasets X3f3 and X5b3 only have two atmcal spectral windows: '''spw 4''' and '''spw 6''') | ||
* '''spw | |||
The final column of the listobs output in the logger | The final column of the listobs output in the logger gives the scan intent. Later we will use this information to flag the pointing scans and the hot and ambient load calibration scans. | ||
We'll now have a look at the configuration of the antennas used to take the data using the task {{plotants}} (<xr id="uid___A002_X5e971a_X124.plotants.png"/>). | We'll now have a look at the configuration of the antennas used to take the data using the task {{plotants}} (<xr id="uid___A002_X5e971a_X124.plotants.png"/>). | ||
| Line 475: | Line 504: | ||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
print "Antenna configuration for : "+ | os.system('rm -rf '+name+'.plotants.png') | ||
plotants(vis= | print "Antenna configuration for : "+name | ||
plotants(vis=name+'.ms', figfile=name+'.plotants.png') | |||
dummy_string = raw_input("Hit <Enter> to see the antenna configuration for the next data set.") | dummy_string = raw_input("Hit <Enter> to see the antenna configuration for the next data set.") | ||
</source> | </source> | ||
This will loop through all 6 data sets, show you the antenna position for each, and save that as a file named, e.g., "uid___A002_X5e971a_X124.plotants.png" for the first data set. The "raw_input" command asks CASA to wait for your input before proceeding. If you would prefer to just browse the .png files after the fact you can remove this. Notice that the antenna setup changes, but only slightly, over the course of the 6 data sets. | This will loop through all 6 data sets, show you the antenna position for each, and save that as a file named, e.g., "uid___A002_X5e971a_X124.plotants.png" for the first data set. The "raw_input" command asks CASA to wait for your input before proceeding. If you would prefer to just browse the .png files after the fact you can remove this. Notice that the antenna setup changes, but only slightly, over the course of the 6 data sets. | ||
==''A Priori'' Flagging== | ==''A Priori'' Flagging== | ||
Even before we look in detail, we know that there are some data that we wish to exclude. We will start by flagging "shadowed" data where one antenna blocks the line of sight of another. We will also flag scans that were used to carry out pointing and atmospheric calibration, identified by their scan intent. Finally, we'll flag the autocorrelation data (the correlation of the signal from an antenna with itself) as we are only interested in cross-correlation data to make an interferometric image. | Even before we look in detail, we know that there are some data that we wish to exclude. We will start by flagging "shadowed" data where one antenna blocks the line of sight of another. We will also flag scans that were used to carry out pointing and atmospheric calibration, identified by their scan intent. Finally, we'll flag the autocorrelation data (the correlation of the signal from an antenna with itself) as we are only interested in cross-correlation data to make an interferometric image. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 512: | Line 524: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in basename: | ||
print " "+ | print " "+name | ||
flagcmd(vis= | flagcmd(vis=name+'.ms', inpmode='table', action='apply') | ||
</source> | </source> | ||
| Line 523: | Line 535: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in basename: | ||
print "Flagging shadowed data for "+asdm | print "Flagging shadowed data for "+asdm | ||
flagdata(vis= | flagdata(vis=name+'.ms',mode = 'shadow', flagbackup = F) | ||
</source> | </source> | ||
In the {{flagdata}} task we choose: | In the {{flagdata}} task we choose: | ||
* '''vis''' = | * '''vis''' = name+'.ms' : each measurement set | ||
* '''mode''' = 'shadow': flag shadowed data | * '''mode''' = 'shadow': flag shadowed data | ||
* '''flagbackup''' = F: Do not automatically back up the flag files. We will save all of the ''a priori'' flags together using {{flagmanager}} at the end of this subsection and save some space and time. | * '''flagbackup''' = F: Do not automatically back up the flag files. We will save all of the ''a priori'' flags together using {{flagmanager}} at the end of this subsection and save some space and time. | ||
| Line 538: | Line 550: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in basename: | ||
print "Flagging calibration scans for "+ | print "Flagging calibration scans for "+name | ||
flagdata(vis= | flagdata(vis=name+'.ms', mode='manual', intent='*POINTING*,*SIDEBAND_RATIO*,*ATMOSPHERE*', flagbackup = F) | ||
</source> | </source> | ||
| Line 549: | Line 561: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in basename: | ||
print "Flagging autocorrelation data for "+ | print "Flagging autocorrelation data for "+name | ||
flagdata(vis= | flagdata(vis=name+'.ms',autocorr=True,flagbackup=F) | ||
</source> | </source> | ||
| Line 558: | Line 570: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in basename: | ||
print "Backing up 'a priori' flags for "+ | print "Backing up 'a priori' flags for "+name | ||
flagmanager(vis = | flagmanager(vis = name+'.ms', mode = 'save', versionname = 'Apriori') | ||
</source> | </source> | ||
| Line 579: | Line 591: | ||
==Create and Apply Tsys and Antenna Position Calibration Tables== | ==Create and Apply Tsys and Antenna Position Calibration Tables== | ||
===Tsys=== | ===Tsys=== | ||
| Line 596: | Line 604: | ||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
os.system('rm -rf '+name+'.ms.tsys') | |||
gencal(vis = name+'.ms', | |||
caltable= | caltable=name+'.ms.tsys', | ||
caltype = 'tsys') | caltype = 'tsys') | ||
au.plotbandpass(caltable=asdm+'.ms.tsys',yaxis='amp',xaxis='freq | |||
for i in range(0,6): | |||
asdm = basename[i] | |||
au.plotbandpass(caltable=asdm+'.ms.tsys', yaxis='amp', xaxis='freq', | |||
buildpdf=False, figfile='./plots_tsys/tsysall'+spw+'_'+asdm+'.png', interactive=False, | |||
antenna='CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12',chanrange='8~120', | |||
subplot=42,overlay='antenna') | |||
</source> | </source> | ||
This sequence loops over all of our files and plots Tsys as a function of time for channel | This sequence loops over all of our files and plots Tsys as a function of time for channel. In the call to {{plotcal}}: | ||
The Tsys values in CM01 and CM07 are too high. They will be flagged. | The Tsys values in CM01 and CM07 are too high. They will be flagged. | ||
* '''subplot'''=42 parameter sets up a 4 x 2 panel grid. | * '''subplot'''=42 parameter sets up a 4 x 2 panel grid. | ||
* '''iteration''' tells {{plotcal}} to make a separate plot for each antenna. | * '''iteration''' tells {{plotcal}} to make a separate plot for each antenna. | ||
The 'raw_input' commands will wait for you to hit Enter before issuing the next plot command. In the example above these are commented out (the leading "#" means that CASA will ignore them). If you would like to interactively cycle through the plots, uncomment them by removing the "#". Otherwise, the '''figfile''' parameter directs the output to .png files for later inspection. The easiest way to look at the 20 plots produced here is to simply inspect the .png files using your favorite viewer. | The 'raw_input' commands will wait for you to hit Enter before issuing the next plot command. In the example above these are commented out (the leading "#" means that CASA will ignore them). If you would like to interactively cycle through the plots, uncomment them by removing the "#". Otherwise, the '''figfile''' parameter directs the output to .png files for later inspection. The easiest way to look at the 20 plots produced here is to simply inspect the .png files using your favorite viewer. | ||
| Line 619: | Line 630: | ||
<figure id="uid___A002_X5e971a_X124.spw8.t0.png"> | <figure id="uid___A002_X5e971a_X124.spw8.t0.png"> | ||
[[File:uid___A002_X5e971a_X124.spw8.t0.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X5e971a_X124 | [[File:uid___A002_X5e971a_X124.spw8.t0.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X5e971a_X124.</caption>]] | ||
</figure> | </figure> | ||
<figure id="uid___A002_X5e971a_X124.spw10.t3.png"> | <figure id="uid___A002_X5e971a_X124.spw10.t3.png"> | ||
[[File:uid___A002_X5e971a_X124.spw10.t3.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X5e971a_X124 | [[File:uid___A002_X5e971a_X124.spw10.t3.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X5e971a_X124. </caption>]] | ||
</figure> | </figure> | ||
<figure id="uid___A002_X5e971a_X124.spw14.t1.png"> | <figure id="uid___A002_X5e971a_X124.spw14.t1.png"> | ||
[[File:uid___A002_X5e971a_X124.spw14.t1.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X5e971a_X124 | [[File:uid___A002_X5e971a_X124.spw14.t1.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X5e971a_X124.</caption>]] | ||
</figure> | </figure> | ||
Now have a look at the Tsys vs. frequency plots or see <xr id="uid___A002_X5e971a_X124.spw8.t0.png"/>, <xr id="uid___A002_X5e971a_X124.spw10.t3.png"/>, and <xr id="uid___A002_X5e971a_X124.spw14.t1.png"/> for examples on the first data set. You can see the effect of a close pair of atmospheric ozone absorption lines at about | Now have a look at the Tsys vs. frequency plots or see <xr id="uid___A002_X5e971a_X124.spw8.t0.png"/>, <xr id="uid___A002_X5e971a_X124.spw10.t3.png"/>, and <xr id="uid___A002_X5e971a_X124.spw14.t1.png"/> for examples on the first data set. You can see the effect of a close pair of atmospheric ozone absorption lines at about 101.8 GHz that makes Tsys larger near that frequency in all antennas. Applying the Tsys calibration tables will minimize the contribution of these atmospheric lines. | ||
Additional issues: | |||
We will flag the spikes, which are appearing in Tsys caltable. | |||
<source lang="python"> | |||
#In CASA | |||
flagdata(vis = 'uid___A002_X60b415_X44.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM09', spw = '8:32', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X60b415_X44.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM09', spw = '8:96', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM02', spw = '10:94~97', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM02', spw = '12:93~97', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM06', spw = '10:96~97', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM06', spw = '12:95~97', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM07', spw = '10:99~101', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys', | |||
mode = 'manual', | |||
antenna = 'CM07', spw = '12:99~101', | |||
flagbackup = T) | |||
</source> | |||
X124 and X2e7 have erratic behavior over parts of Tsys spectral window for spw=10 and 12: applying Tsys 'as is' produces artifacts, visible e.g. in amp vs chan plots for the phase calibrator | |||
(which look reasonable before applying Tsys). | |||
<source lang="python"> | |||
#In CASA | |||
flagdata(vis = 'uid___A002_X5e971a_X124.ms.tsys', | |||
mode = 'manual', | |||
spw = '10:38~90', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X5e971a_X124.ms.tsys', | |||
mode = 'manual', | |||
spw = '12:38~90', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X5e971a_X2e7.ms.tsys', | |||
mode = 'manual', | |||
spw = '10:38~90', | |||
flagbackup = T) | |||
flagdata(vis = 'uid___A002_X5e971a_X2e7.ms.tsys', | |||
mode = 'manual', | |||
spw = '12:38~90', | |||
flagbackup = T) | |||
</source> | |||
===Antenna Positions=== | ===Antenna Positions=== | ||
The antenna position table reflects refinements in the measured positions of the antennas from those stored in the data. gencal will now be used put antenna position data into each observation. Again, gencal will merely append to existing antenna position data, ruining any subsequent results. We start by removing any existing antenna position refinements, followed by defining the antenna names, then their refinements (both as arrays), finally running gencal to create the information CASA can refer to for antenna positions. | The antenna position table reflects refinements in the measured positions of the antennas from those stored in the data. gencal will now be used put antenna position data into each observation. Again, gencal will merely append to existing antenna position data, ruining any subsequent results. We start by removing any existing antenna position refinements, followed by defining the antenna names, then their refinements (both as arrays), finally running gencal to create the information CASA can refer to for antenna positions. | ||
Note:Although four dataset (X124, X2e7, X3f3 and X5b3) have bad parameters of antenna positions, one dataset (X44) has correct parameters of antenna position, so we estimated "parameters" in gencal of four dataset using the antenna position table of X44 as a reference. | |||
X4eb has very good antenna position because the observation was done under good baseline condition (parameter in gencal is zero). Antenna position corrections are not necessary for X4eb. | |||
<source lang="python"> | <source lang="python"> | ||
#In CASA | #In CASA | ||
os.system('rm -rf | os.system('rm -rf *.ms.antpos') | ||
gencal(vis = 'uid___A002_X5e971a_X124.ms', | gencal(vis = 'uid___A002_X5e971a_X124.ms', | ||
| Line 642: | Line 730: | ||
caltype = 'antpos', | caltype = 'antpos', | ||
antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12', | antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12', | ||
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,1.04838982e-05,4.57765535e-04,3.53057869e-04, | parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04, | ||
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,-8.91945325e-04,4.27036546e-04,1.40665658e-03,-1.16256997e-04, | 1.04838982e-05,4.57765535e-04,3.53057869e-04, | ||
2.12460477e-03,-5.08893328e-03,-3.65406508e-03,1.86459431e-02,5.67199755e-03,8.04639747e-03,4.01791865e-02, | 1.38600077e-03,-6.59998506e-04,-4.26100381e-03, | ||
1.46969082e-03,-5.75850718e-05,6.55882061e-04, | -8.91945325e-04,4.27036546e-04,1.40665658e-03, | ||
-1.16256997e-04,2.12460477e-03,-5.08893328e-03, | |||
-3.65406508e-03,1.86459431e-02,5.67199755e-03, | |||
8.04639747e-03,4.01791865e-02,1.46969082e-03, | |||
-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | |||
gencal(vis = 'uid___A002_X5e971a_X2e7.ms', | gencal(vis = 'uid___A002_X5e971a_X2e7.ms', | ||
| Line 656: | Line 743: | ||
caltype = 'antpos', | caltype = 'antpos', | ||
antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12', | antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12', | ||
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,1.04838982e-05,4.57765535e-04,3.53057869e-04, | parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04, | ||
1.38600077e-03,-6.59998506e-04,-4.26100381e-03, -8.91945325e-04,4.27036546e-04,1.40665658e-03,-1.16256997e-04, | 1.04838982e-05,4.57765535e-04,3.53057869e-04, | ||
2.12460477e-03,-5.08893328e-03,-3.65406508e-03,1.86459431e-02,5.67199755e-03,8.04639747e-03,4.01791865e-02, | 1.38600077e-03,-6.59998506e-04,-4.26100381e-03, | ||
1.46969082e-03,-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | -8.91945325e-04,4.27036546e-04,1.40665658e-03, | ||
-1.16256997e-04,2.12460477e-03,-5.08893328e-03, | |||
-3.65406508e-03,1.86459431e-02,5.67199755e-03, | |||
8.04639747e-03,4.01791865e-02,1.46969082e-03, | |||
-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | |||
gencal(vis = 'uid___A002_X5e9ff1_X3f3.ms', | gencal(vis = 'uid___A002_X5e9ff1_X3f3.ms', | ||
| Line 666: | Line 756: | ||
caltype = 'antpos', | caltype = 'antpos', | ||
antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12', | antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12', | ||
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,1.04838982e-05,4.57765535e-04,3.53057869e-04, | parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04, | ||
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,-8.91945325e-04, | 1.04838982e-05,4.57765535e-04,3.53057869e-04, | ||
2.12460477e-03,-5.08893328e-03,-3.65406508e-03,1.86459431e-02,5.67199755e-03,8.04639747e-03,4.01791865e-02, | 1.38600077e-03,-6.59998506e-04,-4.26100381e-03, | ||
1.46969082e-03,-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | -8.91945325e-04,4.27036546e-04,1.40665658e-03, | ||
-1.16256997e-04,2.12460477e-03,-5.08893328e-03, | |||
-3.65406508e-03,1.86459431e-02,5.67199755e-03, | |||
8.04639747e-03,4.01791865e-02,1.46969082e-03, | |||
-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | |||
gencal(vis = 'uid___A002_X5e9ff1_X5b3.ms', | gencal(vis = 'uid___A002_X5e9ff1_X5b3.ms', | ||
caltable = 'uid___A002_X5e9ff1_X5b3.ms.antpos', | caltable = 'uid___A002_X5e9ff1_X5b3.ms.antpos', | ||
caltype = 'antpos', | caltype = 'antpos', | ||
antenna = 'CM02,CM03,CM04,CM05,CM06,CM09,CM12', | antenna = 'CM02,CM03,CM04,CM05,CM06,CM09,CM12', | ||
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,1.04838982e-05,4.57765535e-04,3.53057869e-04, | parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04, | ||
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,-8.91945325e-04,4.27036546e-04,1.40665658e-03,-1.16256997e-04, | 1.04838982e-05,4.57765535e-04,3.53057869e-04, | ||
2.12460477e-03,-5.08893328e-03,8.04639747e-03,4.01791865e-02,1.46969082e-03,-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | 1.38600077e-03,-6.59998506e-04,-4.26100381e-03, | ||
-8.91945325e-04,4.27036546e-04,1.40665658e-03, | |||
-1.16256997e-04,2.12460477e-03,-5.08893328e-03, | |||
8.04639747e-03,4.01791865e-02,1.46969082e-03, | |||
-5.75850718e-05,6.55882061e-04,7.88089819e-05]) | |||
gencal(vis = 'uid___A002_X60b415_X44.ms', | gencal(vis = 'uid___A002_X60b415_X44.ms', | ||
caltable = 'uid___A002_X60b415_X44.ms.antpos', | caltable = 'uid___A002_X60b415_X44.ms.antpos', | ||
caltype = 'antpos', | caltype = 'antpos', | ||
antenna = 'CM12,CM02,CM03,CM05', | antenna = 'CM12,CM02,CM03,CM05', | ||
parameter = [5.70082944295e-05,-0.00065711393962,-7.91980095891e-05,0.000104837426825,-0.000430660050663, | parameter = [5.70082944295e-05,-0.00065711393962,-7.91980095891e-05,0.000104837426825, | ||
-0.000411915081903,-1.1060689293e-05,-0.000458996548728,-0.00035344706733, | -0.000430660050663,-0.000411915081903,-1.1060689293e-05,-0.000458996548728, | ||
-0.00035344706733,0.000982235185802,-0.000412690453231,-0.00145998690277]) | |||
gencal(vis = ' | |||
caltable = ' | gencal(vis = 'uid___A002_X62f759_X4eb.ms', | ||
caltable = 'uid___A002_X62f759_X4eb.ms.antpos', | |||
caltype = 'antpos', | caltype = 'antpos', | ||
antenna = ' | antenna = 'CM09', | ||
parameter = [ | parameter = [0,0,0]) | ||
</source> | </source> | ||
=== | ===Applying antpos and Tsys tables=== | ||
We are now ready to apply the Tsys tables to the data with {{applycal}}, which reads the specified gain calibration tables, applies them to the (raw) data column, and writes the calibrated results into the corrected column. Again, we loop through all the datasets. It is important to only apply Tsys obtained close in time to the data being corrected, so in addition to looping over data sets we define the list of unique source names and loop over these. Then by setting '''gainfield''' and '''field''' to the same value we ensure that Tsys are only applied to the source for which they are measured. | We are now ready to apply the Tsys tables to the data with {{applycal}}, which reads the specified gain calibration tables, applies them to the (raw) data column, and writes the calibrated results into the corrected column. Again, we loop through all the datasets. It is important to only apply Tsys obtained close in time to the data being corrected, so in addition to looping over data sets we define the list of unique source names and loop over these. Then by setting '''gainfield''' and '''field''' to the same value we ensure that Tsys are only applied to the source for which they are measured. | ||
The applycal task now has much more flexibility for interpolating and applying calibrations derived in one spectral window to another, even if they do not share the same spectral shape (number of channels and channel width). This new functionality is used below to interpolate the TDM (128 channel) Tsys measurements to the FDM (4080 channel) spectral windows. This is controlled through the spectral window mapping parameter *spwmap*. Because this can be a bit confusing, we've written a "helper" function that will tell you what you should put for the Tsys calibration table part of spwmap. We only need to run it on one of the datasets because they are all the same in this regard | The applycal task now has much more flexibility for interpolating and applying calibrations derived in one spectral window to another, even if they do not share the same spectral shape (number of channels and channel width). This new functionality is used below to interpolate the TDM (128 channel) Tsys measurements to the FDM (4080 channel) spectral windows. This is controlled through the spectral window mapping parameter *spwmap*. Because this can be a bit confusing, we've written a "helper" function that will tell you what you should put for the Tsys calibration table part of spwmap. We only need to run it on one of the datasets because they are all the same in this regard. | ||
<source lang="python"> | <source lang="python"> | ||
# | # Re-entering our array | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | |||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | |||
</source> | |||
for | Since some datasets have 4 spws and other datasets have 2 spws the application of the calibration tables needs to be treated split into several steps. Also, X5b3 has the Tsys for the science fields done on the phase calibrator (the other datasets have Tsys done on M100 itself) and X44 does not have an observation of Titan as its amplitude calibrator, so we also need to treat these separately. | ||
Now run the applycal commands. | Now run the applycal commands. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | #In CASA | ||
from recipes.almahelpers import tsysspwmap | |||
# When you run tsysspwmap(vis=name+'.ms',tsystable=name+'.ms.tsys') this will print, for example for uid___A002_X5e971a_X124: | |||
# Datasets with 4 spws | |||
for | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | ||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1229+0203', | applycal(vis=name+'.ms', field = 'J1229+0203', | ||
spw='16,18,20,22', | spw='16,18,20,22', | ||
gaintable = [name+'.ms | gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | ||
spwmap = [tsysmap], | spwmap = [tsysmap,[]], | ||
interp = 'linear, linear', | |||
gainfield=['J1229+0203',''], | gainfield=['J1229+0203',''], | ||
calwt=T, | calwt=T, | ||
flagbackup=F) | flagbackup=F) | ||
applycal(vis=name+'.ms', field = ' | # Datasets where the Tsys is done on M100 | ||
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X62f759_X4eb']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1215+1654,M100', | |||
spw='16,18,20,22', | spw='16,18,20,22', | ||
gaintable = [name+'.ms | gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | ||
spwmap = [tsysmap], | spwmap = [tsysmap,[]], | ||
interp = 'linear, linear', | |||
gainfield=['M100'], | |||
calwt=T, | calwt=T, | ||
flagbackup=F) | flagbackup=F) | ||
# Dataset where Tsys is done on the phase calibrator rather than M100 | |||
for name in ['uid___A002_X60b415_X44']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1215+1654,M100', | applycal(vis=name+'.ms', field = 'J1215+1654,M100', | ||
spw='16,18,20,22', | spw='16,18,20,22', | ||
gaintable = [name+'.ms | gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | ||
spwmap = [tsysmap], | spwmap = [tsysmap,[]], | ||
interp = 'linear, linear', | |||
gainfield=['J1215+1654'], | |||
calwt=T, | calwt=T, | ||
flagbackup=F) | flagbackup=F) | ||
# Datasets that have observations of Titan | |||
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'Titan', | |||
spw='16,18,20,22', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='Titan', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
# Datasets with 2 spws | |||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1229+0203', | |||
spw='8,10', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='J1229+0203', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
applycal(vis=name+'.ms', field = 'Titan', | |||
spw='8,10', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='Titan', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
applycal(vis=name+'.ms', field = 'J1215+1654,M100', | |||
spw='8,10', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='M100', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
</source> | </source> | ||
| Line 751: | Line 897: | ||
*'''field''': the field ''to'' which we will apply the calibration, | *'''field''': the field ''to'' which we will apply the calibration, | ||
*'''gainfield''': the field ''from'' which we wish to take the calibration table | *'''gainfield''': the field ''from'' which we wish to take the calibration table | ||
==Inspect Data== | ==Inspect Data== | ||
| Line 871: | Line 934: | ||
First, we plot amplitude as a function of frequency for 3c273. We start by plotting the DATA column, set color to indicate the two correlations (i.e., the XX and YY polarizations), and ask plotms to iterate over baseline. By setting '''antenna''' to 'CM04&*' we select only baselines that include CM04. We ask plotms to average all data over a very long timescale, '''avgtime''' = 1e8 seconds. By setting '''avgscan''' = True we allow {{plotms}} to average across scan boundaries. The result is a plot of average amplitude per channel vs. frequency. | First, we plot amplitude as a function of frequency for 3c273. We start by plotting the DATA column, set color to indicate the two correlations (i.e., the XX and YY polarizations), and ask plotms to iterate over baseline. By setting '''antenna''' to 'CM04&*' we select only baselines that include CM04. We ask plotms to average all data over a very long timescale, '''avgtime''' = 1e8 seconds. By setting '''avgscan''' = True we allow {{plotms}} to average across scan boundaries. The result is a plot of average amplitude per channel vs. frequency. | ||
Here, we will show the dataset with 4BBs(X124, X2e7, X44, and X4eb) and 2BBs(X3f3 and X5b3), separately. | |||
For 4BBs dataset, | |||
<source lang="python"> | |||
# In CASA | |||
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | |||
plotms(vis=name+'.ms', | |||
field='J1229+0203', | |||
xaxis='frequency', yaxis='amp', | |||
selectdata=T, spw='22', | |||
avgtime='1e8',avgscan=T, | |||
coloraxis='corr', | |||
iteraxis='baseline', | |||
antenna='CM04&*', | |||
ydatacolumn='data') | |||
</source> | |||
The rest dataset are here. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
plotms(vis= | plotms(vis=name+'.ms', | ||
field='J1229+0203', | field='J1229+0203', | ||
xaxis='frequency', yaxis='amp', | xaxis='frequency', yaxis='amp', | ||
selectdata=T, spw=' | selectdata=T, spw='10', | ||
avgtime='1e8',avgscan=T, | avgtime='1e8',avgscan=T, | ||
coloraxis='corr', | coloraxis='corr', | ||
| Line 888: | Line 969: | ||
ydatacolumn='data') | ydatacolumn='data') | ||
</source> | </source> | ||
Notice the green arrows along the bottom of the {{plotms}} window. We asked {{plotms}} to iterate over baseline. As you click the arrows, the plot will rotate from baseline to baseline, always with CM04 so that each antenna shows up once. To see the effect of the calibration, go to the "Axes" tab along the left of the {{plotms}} window and pull down the Data Column menu under the Y Axis. Set this from DATA to CORRECTED and you should see the effects of the calibration. You may need to ensure that the "Force Reload" box is checked before clicking "Plot" (both buttons lie at the bottom of the panel). For the most part things get better (flatter). | Notice the green arrows along the bottom of the {{plotms}} window. We asked {{plotms}} to iterate over baseline. As you click the arrows, the plot will rotate from baseline to baseline, always with CM04 so that each antenna shows up once. To see the effect of the calibration, go to the "Axes" tab along the left of the {{plotms}} window and pull down the Data Column menu under the Y Axis. Set this from DATA to CORRECTED and you should see the effects of the calibration. You may need to ensure that the "Force Reload" box is checked before clicking "Plot" (both buttons lie at the bottom of the panel). For the most part things get better (flatter). | ||
<figure id="plotms_amp_vs_time.png"> | <figure id="plotms_amp_vs_time.png"> | ||
[[File:plotms_amp_vs_time.png|200px|thumb|right|<caption> Example of amplitude vs. time | [[File:plotms_amp_vs_time.png|200px|thumb|right|<caption> Example of amplitude vs. time before correction.</caption>]] | ||
</figure> | </figure> | ||
<figure id="plotms_amp_vs_time_corr.png"> | <figure id="plotms_amp_vs_time_corr.png"> | ||
[[File:plotms_amp_vs_time_corr.png|200px|thumb|right|<caption> Example of amplitude vs. time | [[File:plotms_amp_vs_time_corr.png|200px|thumb|right|<caption> Example of amplitude vs. time after correction.</caption>]] | ||
</figure> | </figure> | ||
You can now make analogous calls to examine the phase vs. frequency, amplitude vs. time, and phase vs. time. | You can now make analogous calls to examine the phase vs. frequency, amplitude vs. time, and phase vs. time. | ||
Here, we will show the examples of 4BBs dataset (X124 for this case). | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis= | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
name=basename[0] | |||
plotms(vis=name+'.ms', | |||
field='J1229+0203', | field='J1229+0203', | ||
xaxis='frequency', yaxis='phase', | xaxis='frequency', yaxis='phase', | ||
selectdata=T, spw= | selectdata=T, spw='22', | ||
avgtime='1e8',avgscan=T, | avgtime='1e8',avgscan=T, | ||
coloraxis='corr', | coloraxis='corr', | ||
| Line 912: | Line 997: | ||
ydatacolumn='data') | ydatacolumn='data') | ||
plotms(vis= | plotms(vis=name+'.ms', | ||
field=' | field='', | ||
xaxis='time', yaxis='amp', | xaxis='time', yaxis='amp', | ||
selectdata=T, spw=' | selectdata=T, spw='22:300~3800', | ||
avgchannel=' | avgchannel='4000',avgscan=F, | ||
coloraxis=' | coloraxis='field', | ||
iteraxis=' | iteraxis='', | ||
antenna='CM04&*', | antenna='CM04&*', | ||
ydatacolumn='data') | ydatacolumn='data') | ||
plotms(vis= | plotms(vis=name+'.ms', | ||
field=' | field='', | ||
xaxis='time', yaxis='phase', | xaxis='time', yaxis='phase', | ||
selectdata=T, spw=' | selectdata=T, spw='22:300~3800', | ||
avgchannel=' | avgchannel='4000',avgscan=F, | ||
coloraxis='corr', | coloraxis='corr', | ||
iteraxis='baseline', | iteraxis='baseline', | ||
| Line 935: | Line 1,020: | ||
Where: | Where: | ||
* '''avgchannel''' set to a large number causes the plots of phase and amplitude vs. time to average data at all frequencies into a single point for each measurement. | * '''avgchannel''' set to a large number causes the plots of phase and amplitude vs. time to average data at all frequencies into a single point for each measurement (we exclude the channels at the edge of the spw as for those the Tsys is poorly measured). | ||
* '''coloraxis''' corr sets the colors to correspond to the two polarizations of the data. | * '''coloraxis''' corr sets the colors to correspond to the two polarizations of the data. | ||
| Line 942: | Line 1,027: | ||
This is a lot of data inspection and that's only for one of 6 data sets! You can iterate across the data by hand, updating "asdm" to refer to each data set in order and cycling between baselines and DATA/CORRECTED. It is also possible to script CASA to show you the key plots in succession (see the next block down). However you approach the infrastructure, you are looking for: | This is a lot of data inspection and that's only for one of 6 data sets! You can iterate across the data by hand, updating "asdm" to refer to each data set in order and cycling between baselines and DATA/CORRECTED. It is also possible to script CASA to show you the key plots in succession (see the next block down). However you approach the infrastructure, you are looking for: | ||
* Improved scatter and lower variability in phase and amplitude vs. frequency and time. This indicates that Tsys calibrations helped. | * Improved scatter and lower variability in phase and amplitude vs. frequency and time. This indicates that Tsys (-> amp) and antenna position (-> phase) calibrations helped. | ||
* Sudden jumps in phase or amplitude as a function of either time or frequency. These may indicate problems with the antenna during that observation. | * Sudden jumps in phase or amplitude as a function of either time or frequency. These may indicate problems with the antenna during that observation. | ||
* Large gradients, especially full wraps, in phase as a function of frequency. This may indicate a problem in the delays, the signal path length to the telescopes. | * Large gradients, especially full wraps, in phase as a function of frequency. This may indicate a problem in the delays, the signal path length to the telescopes. | ||
| Line 965: | Line 1,050: | ||
'''Note:''' In loops like this involving {{plotms}}, hitting enter before the last dataset completes loading may cause {{plotms}} to stop refreshing the plot window at each step, or cause the window to disappear altogether. This will be fixed in an upcoming CASA release. Right now, you may need to exit CASA and restart to bring up the {{plotms}} window again, but you should be able to continue in the script where you left off. | '''Note:''' In loops like this involving {{plotms}}, hitting enter before the last dataset completes loading may cause {{plotms}} to stop refreshing the plot window at each step, or cause the window to disappear altogether. This will be fixed in an upcoming CASA release. Right now, you may need to exit CASA and restart to bring up the {{plotms}} window again, but you should be able to continue in the script where you left off. | ||
Here, we will show the examples of 4BBs dataset. | |||
<source lang="python"> | <source lang="python"> | ||
| Line 970: | Line 1,056: | ||
user_input = "" | user_input = "" | ||
for | |||
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | |||
# check if a stop has been requested | |||
if user_input == "stop": | |||
break | |||
# Extract antenna list for this data set. | |||
tb.open(name+'.ms/ANTENNA', nomodify=True) | |||
ants = tb.getcol('NAME') | |||
tb.close | |||
# Define the reference antenna to make baselines with | |||
ref_ant = 'CM04' | |||
# Loop over antennas | |||
for ant in ants: | |||
# Check if the user wants to stop | |||
if user_input == "stop": | |||
break | |||
# Skip correlation of reference antenna with itself (autocorrelations are flagged anyhow) | |||
if ant == ref_ant: | |||
continue | |||
# Define the baseline with the reference antenna for current antenna | |||
ant_str = ref_ant+'&'+ant | |||
print "Showing baseline "+ant_str+" for data set "+name | |||
print "Use this to inspect effect of applying Tsys calibrations." | |||
# Loop over phase and amplitude as axes | |||
for y_axis in ["amp", "phase"]: | |||
# Make 'before' plot for frequency x-axis | |||
print "... "+y_axis+" vs. frequency for DATA:" | |||
plotms(vis=name+'.ms', spw='22', field='J1229+0203',, | |||
antenna=ant_str, xaxis="frequency", yaxis=y_axis, | |||
avgtime="1e8", avgscan=T, coloraxis="corr", | |||
ydatacolumn="data") | |||
user_input = raw_input("Hit <ENTER> to see CORRECTED data [type 'stop'+<Enter> to break out].") | |||
if user_input == "stop": | |||
break | |||
# Make 'after' plot for frequency x-axis | |||
# Make 'before' plot for time x-axis | print "... "+y_axis+" vs. frequency for CORRECTED:" | ||
plotms(vis=name+'.ms', spw='22', field='J1229+0203',, | |||
antenna=ant_str, xaxis="frequency", yaxis=y_axis, | |||
avgtime="1e8", avgscan=T, coloraxis="corr", | |||
ydatacolumn="corrected") | |||
user_input = raw_input("Hit <ENTER> to proceed to next plot [type 'stop'+<Enter> to break out].") | |||
if user_input == "stop": | |||
break | |||
# Make 'before' plot for time x-axis | |||
print "... "+y_axis+" vs. time for DATA:" | |||
plotms(vis=name+'.ms', spw='22', field='J1229+0203',, | |||
antenna=ant_str, xaxis="time", yaxis=y_axis, | |||
avgchannel="1000", coloraxis="corr", | |||
ydatacolumn="data") | |||
user_input = raw_input("Hit <ENTER> to see CORRECTED data [type 'stop'+<Enter> to break out].") | |||
if user_input == "stop": | |||
break | |||
# Make 'after' plot for time y-axis | |||
print "... "+y_axis+" vs. time for CORRECTED:" | |||
plotms(vis=name+'.ms', spw='22', field='J1229+0203',, | |||
antenna=ant_str, xaxis="time", yaxis=y_axis, | |||
avgchannel="1000", coloraxis="corr", | |||
ydatacolumn="corrected") | |||
user_input = raw_input("Hit <ENTER> to proceed to next plot [type 'stop'+<Enter> to break out].") | |||
if user_input == "stop": | |||
break | |||
</source> | </source> | ||
| Line 1,038: | Line 1,134: | ||
A detailed explanation of the procedure is a bit outside the scope of this guide (for more on python see http://www.python.org/ and for more on the CASA toolkit see http://casa.nrao.edu/), but the basic process is to loop over each data set, baseline with the reference antenna (here CM04), and y-axis of interest (phase or amplitude) then plot the effect of the calibration vs. frequency and time for each combination. Running this to step through the data will give you about 200 "before and after" plots from which you could note a subset of problematic cases to be followed up by hand. Many other strategies to inspect the data are also viable. | A detailed explanation of the procedure is a bit outside the scope of this guide (for more on python see http://www.python.org/ and for more on the CASA toolkit see http://casa.nrao.edu/), but the basic process is to loop over each data set, baseline with the reference antenna (here CM04), and y-axis of interest (phase or amplitude) then plot the effect of the calibration vs. frequency and time for each combination. Running this to step through the data will give you about 200 "before and after" plots from which you could note a subset of problematic cases to be followed up by hand. Many other strategies to inspect the data are also viable. | ||
With the Tsys applied successfully and the ''a priori'' flagging taken care of we will now {{split}} out the corrected data. We will keep only the corrected data, specified via '''datacolumn''', and only spectral window 1, which contains the FDM (high spectral resolution) observations of the CO(1-0) line. Setting '''keepflags'''=F tells {{split}} not to carry over any fully flagged rows from the original data set to the new MS. | With the Tsys applied successfully and the ''a priori'' flagging taken care of we will now {{split}} out the corrected data. We will keep only the corrected data, specified via '''datacolumn''', and only spectral window 1, which contains the FDM (high spectral resolution) observations of the CO(1-0) line. Setting '''keepflags'''=F tells {{split}} not to carry over any fully flagged rows from the original data set to the new MS. | ||
| Line 1,063: | Line 1,141: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
plotms(vis= | plotms(vis=name+'.ms', | ||
xaxis='time', yaxis='amp', | xaxis='time', yaxis='amp', | ||
avgchannel='4080',coloraxis='field') | avgchannel='4080',coloraxis='field') | ||
dummy_string = raw_input("Examining amplitude vs. time for "+ | dummy_string = raw_input("Examining amplitude vs. time for "+name+" . Hit <Enter> to proceed.") | ||
</source> | </source> | ||
| Line 1,080: | Line 1,158: | ||
Look through the amplitudes vs. time for each data set (remember that we've already examined the phases vs. time and amplitude vs. time for individual baselines above). | Look through the amplitudes vs. time for each data set (remember that we've already examined the phases vs. time and amplitude vs. time for individual baselines above). | ||
Titan is missing in X4eb dataset, we will skip this dataset. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44']: | ||
plotms(vis= | plotms(vis=name+'.ms', | ||
field='J1229+0203', | field='J1229+0203', | ||
xaxis='uvdist', yaxis='amp', | xaxis='uvdist', yaxis='amp', | ||
avgchannel='4080',coloraxis='corr') | avgchannel='4080',coloraxis='corr') | ||
dummy_string = raw_input("Examining amplitude vs. UV Distance for J1229+0203 for "+ | dummy_string = raw_input("Examining amplitude vs. UV Distance for J1229+0203 for "+name+" . Hit <Enter> to proceed.") | ||
plotms(vis= | plotms(vis=name+'.ms', | ||
field='Titan', | field='Titan', | ||
xaxis='uvdist', yaxis='amp', | xaxis='uvdist', yaxis='amp', | ||
avgchannel='4080',coloraxis='corr') | avgchannel='4080',coloraxis='corr') | ||
dummy_string = raw_input("Examining amplitude vs. UV Distance for Titan for "+ | dummy_string = raw_input("Examining amplitude vs. UV Distance for Titan for "+name+" . Hit <Enter> to proceed [type 'stop'+<Enter> to break out].") | ||
# check if a stop has been requested | # check if a stop has been requested | ||
if dummy_string == "stop": | if dummy_string == "stop": | ||
| Line 1,104: | Line 1,179: | ||
</source> | </source> | ||
For this command notice that we can see the CO(1-0) line in the M100 even before calibration (see <xr id=" | For this command notice that we can see the CO(1-0) line in the M100 even before calibration (see <xr id="plotms_amp_freq_M100.png"/>) shows evidence of a strong line. | ||
This suite of plots (along with the earlier inspection of the Tsys tables) gives us the tools we need to identify problematic data through the data sets. We use this to generate a set of inspection-driven {{flagdata}} commands for each data set. | |||
This suite of plots (along with the earlier inspection of the Tsys tables) gives us the tools we need to identify problematic data through the data sets. We use this to generate a set of inspection-driven {{flagdata}} commands for each data set. We apply these before the bandpass and gain calibration. | We apply these before the bandpass and gain calibration. | ||
==Apply Flagging== | ==Apply Flagging== | ||
| Line 1,116: | Line 1,191: | ||
# In CASA | # In CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | |||
flagdata(vis = | |||
for name in basename: | |||
flagdata(vis = name+'.ms',mode='unflag', flagbackup = F) | |||
</source> | </source> | ||
Remember that we dropped the flagged data when splitting out after the | Remember that we dropped the flagged data when splitting out NO SPLIT SO FAR after the Tsys calibration, so this should not undo your "A Priori" flagging of shadowed data, autocorrelations, etc.. In any case this unflagging step is not necessary during the first iteration. | ||
* '''Edge Channels''' | * '''Edge Channels''' | ||
ALMA's sensitivity decreases near the edge of the baseband and it is often useful to check for a 'roll-off' in sensitivity near the edge of the band. This will appear as a downturn in amplitude as a function of channel near the edge of the window in the uncalibrated data, as a flaring due to increased noise at the spw edges in the calibrated data. | |||
ALMA's sensitivity decreases near the edge of the baseband and it is often useful to check for a 'roll-off' in sensitivity near the edge of the band. This will appear as a downturn in amplitude as a function of channel near the edge of the window in the uncalibrated data, as a flaring due to increased noise at the spw edges in the calibrated data. | |||
<figure id="plotms_CM04&CM06_edge.png"> | <figure id="plotms_CM04&CM06_edge.png"> | ||
| Line 1,136: | Line 1,211: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for name in basename: | |||
flagdata(vis = name+'.ms', mode='manual', spw = '*:0~127,*:3952~4079', flagbackup = F) | |||
</source> | |||
* '''Problematic Tsys measurements''' | |||
Above we noted issues with the Tsys measurements for both CM01 and CM07. '''CM07 only for 2 datasets? see below''' We flag the affected data. | |||
<source lang="python"> | |||
# In CASA | |||
for name in basename: | |||
flagdata(vis= name+'.ms', mode='manual', antenna='CM01', flagbackup=F) | |||
for name in basename: | |||
flagdata(vis= name+'.ms', mode='manual', antenna='CM07', flagbackup=F) | |||
</source> | |||
Flagging of individual dataset: | |||
X4eb : there are spikes in spw=18 and 20 for CM02,CM06,CM07. | |||
<source lang="python"> | |||
# In CASA | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms', | |||
mode = 'manual', | |||
antenna = 'CM02', | |||
spw = '18:3080~3110', | |||
flagbackup=F) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms', | |||
mode = 'manual', | |||
antenna = 'CM02', | |||
spw = '20:3010~3105', | |||
flagbackup=F) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms', | |||
mode = 'manual', | |||
antenna = 'CM06', | |||
spw = '18:3080~3128', | |||
flagbackup=F) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms', | |||
mode = 'manual', | |||
field = 'J1229+0203', | |||
antenna = 'CM06', | |||
spw = '20:3020~3120', | |||
flagbackup=F) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms', | |||
mode = 'manual', | |||
field = 'J1229+0203', | |||
antenna = 'CM07', | |||
spw = '18:3160~3250', | |||
flagbackup=F) | |||
flagdata(vis = 'uid___A002_X62f759_X4eb.ms', | |||
mode = 'manual', | |||
field = 'J1229+0203', | |||
antenna = 'CM07', | |||
spw = '20:3140~3240', | |||
flagbackup=F) | |||
</source> | </source> | ||
Flagging of individual dataset: | |||
X3f3 & X5b3 have low amplitude of CM07 | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | |||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | |||
for | for i in [2,3]: | ||
flagdata(vis= | name = basename[i] | ||
print 'Flagging antenna CM07 on ms '+str(i)+': '+asdm+'.ms' | |||
flagdata(vis = name+'.ms', | |||
mode = 'manual', | |||
antenna = 'CM07', | |||
flagbackup=F) | |||
</source> | </source> | ||
* '''Unreliable Short-Spacing Measurements on Titan''' | * '''Unreliable Short-Spacing Measurements on Titan''' | ||
<figure id="plotms_amp_uvdist_titan.png"> | |||
<figure id=" | [[File:plotms_amp_uvdist_titan.png|200px|thumb|right|<caption> Observed amplitude vs. uv-distance for observations of Titan in the first data set. Note the scatter for low projected antenna separations. | ||
[[File: | |||
We will flag these short-spacing data, which may reflect contamination by Saturn, and use only the more extended baselines for flux calibration.</caption>]] | We will flag these short-spacing data, which may reflect contamination by Saturn, and use only the more extended baselines for flux calibration.</caption>]] | ||
</figure> | </figure> | ||
Saturn may contaminate the short u-v spacings from Titan. In any case these often show significant scatter (<xr id="plotms_amp_uvdist_titan.png"/>), so we flag them. | |||
Saturn may contaminate the short u-v spacings from Titan. In any case these often show significant scatter (<xr id=" | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for name in basename: | |||
for | flagdata(vis=name+'.ms', mode='manual', field='Titan', flagbackup = F) | ||
flagdata(vis= | |||
</source> | </source> | ||
Now that we've applied our flagging, back up the flags as version "User" using {{flagmanager}}: | Now that we've applied our flagging, back up the flags as version "User" using {{flagmanager}}: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44']: | ||
flagmanager(vis= | flagmanager(vis=name+'.ms',mode='save',versionname ='User') | ||
</source> | </source> | ||
| Line 1,197: | Line 1,326: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | ||
plotms(vis= | plotms(vis= name+'.ms', | ||
xaxis='freq', yaxis='phase', spw=' | xaxis='freq', yaxis='phase', spw='22', | ||
field='J1229+0203', correlation='XX', | |||
avgtime='1e8', avgscan=T, antenna='*&*', | |||
coloraxis='baseline') | |||
dummy_string = raw_input("Plotting phase vs. frequency for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].") | |||
# check if a stop has been requested | |||
if dummy_string == "stop": | |||
break | |||
</source> | |||
<source lang="python"> | |||
# In CASA | |||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
plotms(vis= name+'.ms', | |||
xaxis='freq', yaxis='phase', spw='10', | |||
field='J1229+0203', correlation='XX', | field='J1229+0203', correlation='XX', | ||
avgtime='1e8', avgscan=T, antenna='*&*', | avgtime='1e8', avgscan=T, antenna='*&*', | ||
coloraxis='baseline') | coloraxis='baseline') | ||
dummy_string = raw_input("Plotting phase vs. frequency for "+ | dummy_string = raw_input("Plotting phase vs. frequency for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].") | ||
# check if a stop has been requested | # check if a stop has been requested | ||
if dummy_string == "stop": | if dummy_string == "stop": | ||
| Line 1,223: | Line 1,365: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
for | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | ||
plotms(vis= | plotms(vis= name+'.ms', | ||
xaxis='time', yaxis='phase', | |||
selectdata=True, field='J1229+0203', | |||
spw='22', antenna='*&*',correlation='XX', | |||
avgchannel='1000', | |||
coloraxis='baseline') | |||
dummy_string = raw_input("Plotting phase vs. time for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].") | |||
# check if a stop has been requested | |||
if dummy_string == "stop": | |||
break | |||
</source> | |||
<source lang="python"> | |||
# In CASA | |||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
plotms(vis= name+'.ms', | |||
xaxis='time', yaxis='phase', | xaxis='time', yaxis='phase', | ||
selectdata=True, field='J1229+0203', | selectdata=True, field='J1229+0203', | ||
spw=' | spw='10', antenna='*&*',correlation='XX', | ||
avgchannel='1000', | avgchannel='1000', | ||
coloraxis='baseline') | coloraxis='baseline') | ||
dummy_string = raw_input("Plotting phase vs. time for "+ | dummy_string = raw_input("Plotting phase vs. time for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].") | ||
# check if a stop has been requested | # check if a stop has been requested | ||
if dummy_string == "stop": | if dummy_string == "stop": | ||
| Line 1,236: | Line 1,394: | ||
</source> | </source> | ||
<xr id="plotms_phase_time_3c273.png"/> shows that the phase varies with time. We need to take this temporal variation into account when we solve for the frequency variations. Therefore we carry out the bandpass calibration in two steps. First, we use {{gaincal}} to solve for the variation of phase as a function of time for 3c273(J1229+0203) on very short timescales. We set {{gaincal}} to derive a separate phase solution for each antenna every integration by setting '''solint''' to 'int'. We solve, averaging together only a small fraction of the total bandpass | <xr id="plotms_phase_time_3c273.png"/> shows that the phase varies with time. We need to take this temporal variation into account when we solve for the frequency variations. Therefore we carry out the bandpass calibration in two steps. First, we use {{gaincal}} to solve for the variation of phase as a function of time for 3c273(J1229+0203) on very short timescales. We set {{gaincal}} to derive a separate phase solution for each antenna every integration by setting '''solint''' to 'int'. We solve, averaging together only a small fraction of the total bandpass to avoid the effects of the phase vs. frequency behavior. We will then apply this solution to remove time-dependent behavior when we solve for the frequency response of the antennas with {{bandpass}}. | ||
<pre style="background-color: #E0FFFF;"> | <pre style="background-color: #E0FFFF;"> | ||
| Line 1,245: | Line 1,403: | ||
<source lang="python"> | <source lang="python"> | ||
#In CASA | #In CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | |||
for | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
print "Running a short solution interval phase calibration for "+ | |||
os.system('rm -rf '+ | for name in basename: | ||
gaincal(vis = | print "Running a short solution interval phase calibration for "+name | ||
os.system('rm -rf '+name+'.bpphase.gcal') | |||
gaincal(vis = name+'.ms', | |||
selectdata=T,field = 'J1229+0203',spw = '', | selectdata=T,field = 'J1229+0203',spw = '', | ||
caltable = | caltable = name+'.bpphase.gcal', | ||
solint = '30.3s',refant = 'CM04', | solint = '30.3s',refant = 'CM04', minsnr = 2.0, minblperant = 3) | ||
</source> | </source> | ||
| Line 1,258: | Line 1,418: | ||
<source lang="python"> | <source lang="python"> | ||
for | for name in basename: | ||
print "Running a bandpass calibration for "+ | print "Running a bandpass calibration for "+name | ||
os.system('rm -rf '+ | os.system('rm -rf '+name+'.bandpass.bcal') | ||
bandpass(vis = | bandpass(vis = name+'.ms', | ||
field = 'J1229+0203', | field = 'J1229+0203', | ||
gaintable = | gaintable = name+'.bpphase.gcal', | ||
caltable = | caltable = name+'.bandpass.bcal', | ||
bandtype='B', | bandtype='B', | ||
solint = 'inf, 16ch',combine = 'scan,field', solnorm=T,refant = 'CM04', | solint = 'inf, 16ch',combine = 'scan,field', solnorm=T,refant = 'CM04', | ||
minsnr=1.0, minblperant = 2) | |||
</source> | </source> | ||
| Line 1,275: | Line 1,435: | ||
*'''gaintable''' specifies any calibration tables to be applied "on the fly" to the data before solving. | *'''gaintable''' specifies any calibration tables to be applied "on the fly" to the data before solving. | ||
*'''solint''' sets the time interval for which solutions are derived for each antenna. | *'''solint''' sets the time interval for which solutions are derived for each antenna. | ||
*'''refant''' set the reference antenna, here | *'''refant''' set the reference antenna, here CM04, which is defined to have zero correction by construction. | ||
*'''calmode''' 'p' sets {{gaincal}} to calibrate the phase only, other options are 'a'mplitude or 'ap' - amplitude and phase. | *'''calmode''' 'p' sets {{gaincal}} to calibrate the phase only, other options are 'a'mplitude or 'ap' - amplitude and phase. | ||
*'''minblperant''' sets the minimum baselines required per antenna for a successful solution. | *'''minblperant''' sets the minimum baselines required per antenna for a successful solution. | ||
| Line 1,295: | Line 1,455: | ||
<figure id="bandpass.CM02.png"> | <figure id="bandpass.CM02.png"> | ||
[[File:bandpass.CM02.png|200px|thumb|right|<caption> Bandpass amplitude ad phase solution for | [[File:bandpass.CM02.png|200px|thumb|right|<caption> Bandpass amplitude ad phase solution for CM02 and the first data set.</caption>]] | ||
</figure> | </figure> | ||
| Line 1,303: | Line 1,463: | ||
<figure id="bandpass_freq_phase.png"> | <figure id="bandpass_freq_phase.png"> | ||
[[File:bandpass_freq_phase.png|200px|thumb|right|<caption> Bandpass phase solution for the | [[File:bandpass_freq_phase.png|200px|thumb|right|<caption> Bandpass phase solution for the rest antennas and the first data set.</caption>]] | ||
</figure> | </figure> | ||
<source lang="python"> | <source lang="python"> | ||
#In CASA | #In CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
os.system('rm -rf cal_plots/*bandpass*.png') | os.system('rm -rf cal_plots/*bandpass*.png') | ||
for | for name in basename: | ||
aU.plotbandpass(caltable= | aU.plotbandpass(caltable=name+'.bandpass.bcal', | ||
field='',xaxis='freq',yaxis='both', | field='',xaxis='freq',yaxis='both', | ||
figfile='cal_plots/'+ | figfile='cal_plots/'+name+'.bandpass.png', | ||
showatm=True,interactive=False,subplot=42) | showatm=True,interactive=False,subplot=42) | ||
</source> | </source> | ||
| Line 1,322: | Line 1,482: | ||
The bandpass calibration will account for the phase and amplitude response of our antennas as a function of frequency. We now solve for the absolute flux scale of the data by referencing to 3c273 and in the next section we will calibrate the phase and amplitude behavior of the antennas as a function of time. However, Titan is not available for these dataset, instead we will use flux of 3c273. | The bandpass calibration will account for the phase and amplitude response of our antennas as a function of frequency. We now solve for the absolute flux scale of the data by referencing to 3c273 and in the next section we will calibrate the phase and amplitude behavior of the antennas as a function of time. However, Titan is not available for these dataset, instead we will use flux of 3c273. | ||
Next, we will use the {{setjy}} task to read the predicted complex visibilities for J1229+0203 into the MODEL column of each data set. | |||
We will put in best flux density for 3c273 instead of Titan. | |||
*X124, X3f3 and X5b3: we will use the value observed on March 17, 2013. | |||
*X2e7 and X44: we will use the value on April 14, 2013. | |||
*X4eb: the latest value will be used. | |||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
# Put in best flux density for 3c273 on Mar 17, 2013 | |||
setjy(vis = 'uid___A002_X5e971a_X124.ms', | |||
field = 'J1229+0203', | |||
fluxdensity = [8.0,0,0,0], | |||
spw = '16,18', | |||
usescratch = F) | |||
setjy(vis = 'uid___A002_X5e971a_X124.ms', | |||
field = 'J1229+0203', | |||
fluxdensity = [7.7,0,0,0], | |||
spw = '20,22', | |||
usescratch = F) | |||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
setjy(vis = name+'.ms', | |||
field = 'J1229+0203', | |||
fluxdensity = [7.7,0,0,0], | |||
spw = '8,10', | |||
usescratch = F) | |||
# Put in best flux density for 3c273 on Apr 14, 2013 | |||
# | for name in ['uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44']: | ||
setjy(vis = name+'.ms', | |||
field = 'J1229+0203', | |||
setjy(vis = | fluxdensity = [8.0,0,0,0], | ||
spw = '16,18', | |||
usescratch = F) | |||
setjy(vis = name+'.ms', | |||
field = 'J1229+0203', | |||
fluxdensity = [7.7,0,0,0], | |||
setjy(vis = | spw = '20,22', | ||
usescratch = F) | |||
# The latest flux density of 3c273 | |||
setjy(vis = 'uid___A002_X62f759_X4eb.ms', | |||
field = 'J1229+0203', | |||
spw = '16,18', | |||
fluxdensity = [7.8,0,0,0], | |||
usescratch = F) | |||
setjy(vis = 'uid___A002_X62f759_X4eb.ms', | |||
field = 'J1229+0203', | |||
spw = '20,22', | |||
fluxdensity = [7.4,0,0,0], | |||
usescratch = F) | |||
</source> | </source> | ||
{{setjy}} will output the flux of 3c273 to the CASA logger and it is worth recording this information. | {{setjy}} will output the flux of 3c273 to the CASA logger and it is worth recording this information. | ||
==Gain (Phase and Amplitude) Calibration== | ==Gain (Phase and Amplitude) Calibration== | ||
| Line 1,367: | Line 1,549: | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
print "Carrying out short timescale phase solution for "+ | print "Carrying out short timescale phase solution for "+name | ||
os.system('rm -rf '+ | os.system('rm -rf '+name+'.intphase.gcal') | ||
gaincal(vis= | gaincal(vis=name+'.ms', | ||
gaintable= | gaintable=name+'.bandpass.bcal', | ||
caltable= | caltable=name+'.intphase.gcal', | ||
calmode='p', | calmode='p', | ||
field='J1229+0203', | field='J1229+0203', | ||
| Line 1,386: | Line 1,568: | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
print "Carrying out longer timescale phase solution for "+ | print "Carrying out longer timescale phase solution for "+name | ||
os.system('rm -rf '+ | os.system('rm -rf '+name+'.scanphase.gcal') | ||
gaincal(vis= | gaincal(vis=name+'.ms', | ||
gaintable= | gaintable=name+'.bandpass.bcal', | ||
caltable= | caltable=name+'.scanphase.gcal', | ||
calmode='p', | calmode='p', | ||
field='J1229+0203', | field='J1229+0203', | ||
| Line 1,405: | Line 1,587: | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
print "Solving for longer (scan) interval amplitude solution for "+ | print "Solving for longer (scan) interval amplitude solution for "+name | ||
os.system('rm -rf '+ | os.system('rm -rf '+name+'.amp.cal') | ||
gaincal(vis = | gaincal(vis = name+'.ms', | ||
gaintable =[ | gaintable =[name+'.bandpass.bcal',name+'.intphase.gcal'], | ||
caltable = | caltable = name+'.amp.cal', | ||
calmode='ap', | calmode='ap', | ||
field = ' | field = ['J1229+0203', 'J1215+1654'], | ||
spw='', | spw='', | ||
refant = 'CM04',solint = 'inf', minsnr=2.0) | refant = 'CM04',solint = 'inf', minsnr=2.0) | ||
</source> | </source> | ||
This "amp.cal" solution gives us the amplitude variations as a function of time, but they are not yet pinned to a realistic scale except in the case of Titan, where we have solved using the model input by {{setjy}}. | This "amp.cal" solution gives us the amplitude variations as a function of time, but they are not yet pinned to a realistic scale except in the case of Titan, where we have solved using the model input by {{setjy}}. | ||
Here, we will set the flux of our secondary calibrator J1215+1654 with reference to 3c273 using {{fluxscale}}. | |||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename: | ||
print "Scaling amplitude calibration to match 3c273 for "+ | print "Scaling amplitude calibration to match 3c273 for "+name | ||
os.system('rm -rf '+ | os.system('rm -rf '+name+'.flux.cal') | ||
fluxscale(vis = | fluxscale(vis = name+'.ms', | ||
caltable = | caltable = name+'.amp.cal', | ||
fluxtable = | fluxtable = name+'.flux.cal', | ||
reference = 'J1229+0203', | reference = 'J1229+0203', | ||
transfer = 'J1215+1654') | transfer = 'J1215+1654') | ||
| Line 1,438: | Line 1,621: | ||
This new correctly-scaled flux table ".flux.cal" replaces the previous ".amp.cal" table as the correct amplitude calibration table to apply to the data, i.e., the ".flux.cal" contains both the time variability of the amplitude solved for in ".amp.cal" and the correct flux scaling set with {{fluxscale}}. | This new correctly-scaled flux table ".flux.cal" replaces the previous ".amp.cal" table as the correct amplitude calibration table to apply to the data, i.e., the ".flux.cal" contains both the time variability of the amplitude solved for in ".amp.cal" and the correct flux scaling set with {{fluxscale}}. | ||
Now we plot the final phase and amplitude calibration tables for each data set. A well-behaved calibration table will show smooth variations as a function of time. Sudden jumps or wild variations among the antenna amplitude gains should prompt further investigation and possibly additional flagging. | Now we plot the final phase and amplitude calibration tables for each data set. A well-behaved calibration table will show smooth variations as a function of time. Sudden jumps or wild variations among the antenna amplitude gains should prompt further investigation and possibly additional flagging. | ||
Here, we will present the examples of X124 dataset. | |||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename[0]: | ||
print "Plotting solutions for "+ | print "Plotting solutions for "+name | ||
plotcal(caltable = | plotcal(caltable = name+'.scanphase.gcal', | ||
xaxis = 'time', yaxis = 'phase', | xaxis = 'time', yaxis = 'phase', | ||
iteration = 'antenna', plotrange=[0,0,-180,180], | iteration = 'antenna', plotrange=[0,0,-180,180], | ||
showgui=False, subplot=421, figfile= | showgui=False, subplot=421, figfile=name+'.scanphase.page1.png', | ||
antenna='', fontsize=6.0) | antenna='', fontsize=6.0) | ||
# dummy_string = raw_input("Hit <Enter> to see next plot.") | # dummy_string = raw_input("Hit <Enter> to see next plot.") | ||
plotcal(caltable = | plotcal(caltable = name+'.scanphase.gcal', | ||
xaxis = 'time', yaxis = 'phase', | xaxis = 'time', yaxis = 'phase', | ||
iteration = 'antenna', plotrange=[0,0,-180,180], | iteration = 'antenna', plotrange=[0,0,-180,180], | ||
showgui=False, subplot=421, figfile= | showgui=False, subplot=421, figfile=name+'.scanphase.page2.png', | ||
antenna='', fontsize=6.0) | antenna='', fontsize=6.0) | ||
# dummy_string = raw_input("Hit <Enter> to see next plot.") | # dummy_string = raw_input("Hit <Enter> to see next plot.") | ||
plotcal(caltable = | plotcal(caltable = name+'.flux.cal', | ||
xaxis = 'time',yaxis = 'amp', | xaxis = 'time',yaxis = 'amp', | ||
plotrange = [0,0,0,0], | plotrange = [0,0,0,0], | ||
antenna='0~7', iteration='antenna', | antenna='0~7', iteration='antenna', | ||
showgui=False, subplot=421, figfile= | showgui=False, subplot=421, figfile=name+'.flux.page1.png', | ||
fontsize=6.0) | fontsize=6.0) | ||
# dummy_string = raw_input("Hit <Enter> to see next plot.") | # dummy_string = raw_input("Hit <Enter> to see next plot.") | ||
plotcal(caltable = | plotcal(caltable = name+'.flux.cal', | ||
xaxis = 'time',yaxis = 'amp', | xaxis = 'time',yaxis = 'amp', | ||
plotrange = [0,0,0,0], | plotrange = [0,0,0,0], | ||
antenna='', iteration='antenna', fontsize=6.0, | antenna='', iteration='antenna', fontsize=6.0, | ||
showgui=False, subplot=421, figfile= | showgui=False, subplot=421, figfile=name+'.flux.page2.png') | ||
# dummy_string = raw_input("Hit <Enter> to see next plot.") | # dummy_string = raw_input("Hit <Enter> to see next plot.") | ||
</source> | </source> | ||
==Apply the Calibrations and Inspect == | ==Apply the Calibrations and Inspect == | ||
Now we will use {{applycal}} to apply the bandpass, phase, and amplitude calibration tables that we generated in the previous sections to the data. We apply the solutions | Now we will use {{applycal}} to apply the bandpass, phase, and amplitude calibration tables that we generated in the previous sections to the data. We apply the solutions to the target source. | ||
<source lang="python"> | <source lang="python"> | ||
#In CASA | #In CASA | ||
from recipes.almahelpers import tsysspwmap | |||
'uid___A002_X60b415_X44',' | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | ||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1229+0203', | |||
spw='16,18,20,22', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
interp = 'linear, linear', | |||
gainfield=['J1229+0203',''], | |||
calwt=T, | |||
flagbackup=F) | |||
# Datasets where the Tsys is done on M100 | |||
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X62f759_X4eb']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1215+1654,M100', | |||
spw='16,18,20,22', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
interp = 'linear, linear', | |||
gainfield=['M100'], | |||
calwt=T, | |||
flagbackup=F) | |||
# Dataset where Tsys is done on the phase calibrator rather than M100 | |||
for name in ['uid___A002_X60b415_X44']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1215+1654,M100', | |||
spw='16,18,20,22', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
interp = 'linear, linear', | |||
gainfield=['J1215+1654'], | |||
calwt=T, | |||
flagbackup=F) | |||
for | # Datasets that have observations of Titan | ||
applycal(vis= | for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44']: | ||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'Titan', | |||
spw='16,18,20,22', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='Titan', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
# Datasets with 2 spws | |||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys') | |||
applycal(vis=name+'.ms', field = 'J1229+0203', | |||
spw='8,10', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='J1229+0203', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
applycal(vis=name+'.ms', field = 'Titan', | |||
spw='8,10', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='Titan', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
applycal(vis=name+'.ms', field = 'J1215+1654,M100', | |||
spw='8,10', | |||
gaintable = [name+'.ms.tsys',name+'.ms.antpos'], | |||
spwmap = [tsysmap,[]], | |||
gainfield='M100', | |||
interp='linear,linear', | |||
calwt=T, | |||
flagbackup=F) | |||
</source> | </source> | ||
| Line 1,501: | Line 1,746: | ||
* '''gaintable''' specifies the tables to apply to the data. There may be several tables applied all at once, here we apply the bandpass, phase, and flux calibrations. | * '''gaintable''' specifies the tables to apply to the data. There may be several tables applied all at once, here we apply the bandpass, phase, and flux calibrations. | ||
* '''interp''' specifies the type of interpolation used to match calibration tables to the data. If there are several gaintables then interp may be either a single value (e.g., "linear" or "nearest") or a list of values with size equal to the list of gaintables. In the case where interp is a list, applycal will match each element to the corresponding gaintable (e.g., first gaintable with first interp parameter). | * '''interp''' specifies the type of interpolation used to match calibration tables to the data. If there are several gaintables then interp may be either a single value (e.g., "linear" or "nearest") or a list of values with size equal to the list of gaintables. In the case where interp is a list, applycal will match each element to the corresponding gaintable (e.g., first gaintable with first interp parameter). | ||
* '''gainfield''' specifies field from which to draw the calibration. As with interp, this can be a list of size matched to gaintable, so that the first element of gainfield will be the field used for the first table | * '''gainfield''' specifies field from which to draw the calibration. As with interp, this can be a list of size matched to gaintable, so that the first element of gainfield will be the field used for the first table. | ||
* '''calwt''' indicates that the weights, which were derived from the Tsys measurements, should also be calibrated. Note that '''calwt''' is True by default. | * '''calwt''' indicates that the weights, which were derived from the Tsys measurements, should also be calibrated. Note that '''calwt''' is True by default. | ||
Once calibrations are applied, it is important to go back and inspect the calibrated data. New problematic antennas or baselines may be visible after calibration. Repeat the steps above, focusing on the CORRECTED data column. Bear in mind that for any point source calibrators we now expect to find phase scattering around zero and to find flat amplitudes as a function of u-v distance. Look for outliers and other signatures of problematic data. As a general rule, you will want to incorporate these data into your overall flagging script then rerun the whole calibration process, so that reduction is iterative. If the data only represent a minor problem, however, it may not be terribly harmful to flag them after the fact so that they do not interfere with imaging but trust that the calibrations are mostly unaffected. | Once calibrations are applied, it is important to go back and inspect the calibrated data. New problematic antennas or baselines may be visible after calibration. Repeat the steps above, focusing on the CORRECTED data column. Bear in mind that for any point source calibrators we now expect to find phase scattering around zero and to find flat amplitudes as a function of u-v distance. Look for outliers and other signatures of problematic data. As a general rule, you will want to incorporate these data into your overall flagging script then rerun the whole calibration process, so that reduction is iterative. If the data only represent a minor problem, however, it may not be terribly harmful to flag them after the fact so that they do not interfere with imaging but trust that the calibrations are mostly unaffected. | ||
<figure id=".png"> | <figure id="plotms_amp_time_phasecal.png"> | ||
[[File: | [[File:plotms_amp_time_phasecal.png|200px|thumb|right|<caption> Calibrated amplitude vs. time plot for J1215+1654 (uid___A002_X5e971a_X124 dataset). </caption>]] | ||
</figure> | </figure> | ||
Here, we will present the examples of X124 dataset. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', | basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', | ||
'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for name in basename[0] : | |||
for | print "Examining calibrated data for "+name | ||
print "Examining calibrated data for "+ | plotms(vis = name+'.ms', xaxis='uvdist', yaxis='amp', | ||
plotms(vis = | ydatacolumn='corrected', field='J1215+1654', | ||
ydatacolumn='corrected', field=' | |||
averagedata=True, avgchannel='4080', avgtime='', | averagedata=True, avgchannel='4080', avgtime='', | ||
avgscan=F, avgbaseline=F, coloraxis='corr') | avgscan=F, avgbaseline=F, coloraxis='corr') | ||
dummy_string = raw_input("Hit <Enter> for next plot.") | dummy_string = raw_input("Hit <Enter> for next plot.") | ||
plotms(vis = | plotms(vis = name+'.ms', xaxis='time', yaxis='amp', | ||
ydatacolumn='corrected', field=' | ydatacolumn='corrected', field='J1215+1654', | ||
averagedata=True, avgchannel='4080', avgtime='', | averagedata=True, avgchannel='4080', avgtime='', | ||
avgscan=F, avgbaseline=F, coloraxis='corr') | avgscan=F, avgbaseline=F, coloraxis='corr') | ||
dummy_string = raw_input("Hit <Enter> for next plot.") | dummy_string = raw_input("Hit <Enter> for next plot.") | ||
plotms(vis = | plotms(vis = name+'.ms', xaxis='freq', yaxis='amp', | ||
ydatacolumn='corrected', field=' | ydatacolumn='corrected', field='J1215+1654',avgtime='1e8', | ||
avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr') | avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr') | ||
dummy_string = raw_input("Hit <Enter> for next plot.") | dummy_string = raw_input("Hit <Enter> for next plot.") | ||
plotms(vis = | plotms(vis = name+'.ms', xaxis='time', yaxis='phase', | ||
ydatacolumn='corrected', field=' | ydatacolumn='corrected', field='J1215+1654', | ||
avgchannel='4080', avgscan=F, avgbaseline=F, coloraxis='corr') | avgchannel='4080', avgscan=F, avgbaseline=F, coloraxis='corr') | ||
dummy_string = raw_input("Hit <Enter> for next plot [type 'stop'+<Enter> to break out].") | dummy_string = raw_input("Hit <Enter> for next plot [type 'stop'+<Enter> to break out].") | ||
| Line 1,543: | Line 1,786: | ||
break | break | ||
</source> | </source> | ||
In <xr id=".png"/> | In <xr id="plotms_amp_time_phasecal.png"/>, we plot phase vs. channel and amp vs. time for J1215+1654 for the X124 dataset. | ||
Finally we can use {{plotms}} to examine the corrected amplitude and phase of | Finally we can use {{plotms}} to examine the corrected amplitude and phase of M100 as a function of time and uv-distance (plots not shown): | ||
<source lang="python"> | <source lang="python"> | ||
| Line 1,574: | Line 1,797: | ||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for | for name in basename[0]: | ||
print "Examining calibrated data for "+ | print "Examining calibrated data for "+name | ||
plotms(vis = | plotms(vis = name+'.split.ms', xaxis='uvdist', yaxis='amp', | ||
ydatacolumn='corrected', field='M100', | ydatacolumn='corrected', field='M100', | ||
averagedata=True, avgchannel='4080', avgtime='', | averagedata=True, avgchannel='4080', avgtime='', | ||
avgscan=F, avgbaseline=F, coloraxis='corr') | avgscan=F, avgbaseline=F, coloraxis='corr') | ||
dummy_string = raw_input("Hit <Enter> for next plot.") | dummy_string = raw_input("Hit <Enter> for next plot.") | ||
plotms(vis = | plotms(vis = name+'.ms', xaxis='freq', yaxis='amp', | ||
ydatacolumn='corrected', field='M100', | ydatacolumn='corrected', field='M100', | ||
avgtime='1e8', avgscan=T, avgbaseline=F, coloraxis='corr') | avgtime='1e8', avgscan=T, avgbaseline=F, coloraxis='corr') | ||
| Line 1,588: | Line 1,811: | ||
==Split and Concatenate Data for M100 Mosaic== | ==Split and Concatenate Data for M100 Mosaic== | ||
The individual data sets are now calibrated. We can safely split out the calibrated data for our science target and drop the calibrators | The individual data sets are now calibrated. We can safely split out the calibrated data for our science target and drop the calibrators. | ||
<source lang="python"> | <source lang="python"> | ||
#In CASA | #In CASA | ||
os.system('rm -rf *.ms.split') | |||
' | |||
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']: | |||
for | split(vis = name+'.ms', | ||
outputvis = name+'.ms.split', | |||
datacolumn = 'corrected', | |||
spw = '16,18,20,22', | |||
keepflags = F) | |||
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']: | |||
split(vis = name+'.ms', | |||
outputvis = name+'.ms.split', | |||
datacolumn = 'corrected', | |||
spw = '8,10', | |||
keepflags = F) | |||
</source> | |||
Before we do this, we will take a brief aside to remove Pointing information from the calibrated data. | Before we do this, we will take a brief aside to remove Pointing information from the calibrated data. | ||
| Line 1,610: | Line 1,843: | ||
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] | ||
for name in basename: | |||
for | print "Manually removing pointing table from "+name+".cal.ms ... Please be careful!" | ||
print "Manually removing pointing table from "+ | tb.open(name+'.cal.ms/POINTING',nomodify=False) | ||
tb.open( | |||
a = tb.rownumbers() | a = tb.rownumbers() | ||
tb.removerows(a) | tb.removerows(a) | ||
| Line 1,622: | Line 1,854: | ||
# In CASA | # In CASA | ||
os.system('rm -rf | os.system('rm -rf M100_Band3_7m_CalibratedData.ms') | ||
concat(vis=['X2e7-cal.ms','X3f3-cal.ms','X4eb-cal.ms','X5b3-cal.ms','X44-cal.ms','X124-cal.ms'],concatvis='M100.7m. | concat(vis=['X2e7-cal.ms','X3f3-cal.ms','X4eb-cal.ms','X5b3-cal.ms','X44-cal.ms','X124-cal.ms'],concatvis='M100_Band3_7m_CalibratedData.ms') | ||
</source> | |||
== Imaging Mosaics == | |||
<pre style="background-color: #E0FFFF;"> | |||
If you are unfamiliar with the basic concepts of deconvolution and clean, pause here and | |||
review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/wilner_synthesis10.pdf | |||
</pre> | |||
Mosaics like other kinds of images are created in the CASA task {{clean}}. To invoke mosaic mode, you simply set the parameter '''imagermode='mosaic''''. The default subparameter '''ftmachine='mosaic'''' is then automatically set. This is a joint deconvolution algorithm that works in the uv-plane. A convolution of the primary beam patterns for each pointing in the mosaic is created: the primary beam response function. The corresponding image of the mosaic response function will be called ''<imagename>.flux.pbcoverage'' and ''<imagename>.flux'' (where the latter differs from the former only if the sensitivity of each field in the mosaic varies). | |||
<pre style="background-color: #E0FFFF;"> | |||
If you want to learn more about mosaicing, pause here and | |||
review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/jott-mosaicking-school-04.pdf | |||
</pre> | |||
==Continuum subtraction and Split out CO(1-0) spw== | |||
In these data, the continuum emission is too weak to contaminate the line emission (i.e. the peak continuum emission is less than the rms noise in the spectral line channels). | |||
Nevertheless, for illustrative purposes we demonstrate how to subtract the continuum emission in the uv-domain using the task {{uvcontsub2}}. | |||
<source lang="python"> | |||
# In CASA | |||
uvcontsub2(vis='M100_Band3_7m_CalibratedData.ms',fitspw='0;1:128~1850;2150~3951',fitorder = 1, combine = 'spw', spw = '0,1') | |||
</source> | |||
Here, '''fitspw''' gives the line-free channels for each mosaic and '''fitorder=1'''. The output MS will have ''.contsub'' appended to the name. | |||
<source lang="python"> | |||
# In CASA | |||
split(vis = 'M100_Band3_7m_CalibratedData.ms', outputvis= 'M100_7m_CO.ms', field='M100',spw='1,3', datacolumn = 'data', keepflags=F) | |||
</source> | |||
== CO(1-0) Imaging == | |||
<figure id="M100_CO1_0_vel.png"> | |||
[[Image:M100_CO1_0_vel.png|thumb|<caption>M100-7m CO(1-0) uv-spectrum in LSRK velocity space.</caption>]] | |||
</figure> | |||
Now we are ready to make cubes of the line emission. The imaging parameters are similar to the continuum except for | |||
those dealing with the spectral setup: '''mode''', '''start''', '''width''', '''nchan''', '''restfreq''', and '''outframe''' parameters. When making spectral images you have three choices for the '''mode''' parameter: '''channel''', '''velocity''', and '''frequency'''. Data are taken using constant frequency channels. For spectral line analysis it's often more useful to have constant velocity channels, and this is also the best way to make images of multiple lines with the exact same channelization for later comparison. For '''mode='velocity'''', the desired '''start''' and '''width''' also need to be given in velocity units for the desired output frame. | |||
It is important to note that ALMA does not do on-line Doppler Tracking and the native frame of the data is TOPO. If you do not specify '''outframe''' the output cube will also be in TOPO, which is not very useful for scientific analysis. The Doppler Shift is taken out during the regridding to the desired outframe in {{clean}} or alternatively it can be done separately by the {{cvel}} task which would need to be run before {{clean}}. | |||
At first, we will make initial dirty image. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf M100.CO1_0_vel.png') | |||
plotms(vis='M100_Band3_7m_CalibratedData.ms.contsub',xaxis='velocity',yaxis='amp', | |||
avgtime='1e8',avgscan=T,transform=T,freqframe='LSRK', | |||
restfreq='115.271201800GHz',plotfile='M100.CO1_0_vel.png') | |||
</source> | |||
<source lang="python"> | |||
# In CASA | |||
# Define clean parameters | |||
vis='M100_7m_CO.ms' | |||
prename='M100_7m_cube' | |||
myimage=prename+'.image' | |||
myflux=prename+'.flux' | |||
mymask=prename+'.mask' | |||
myresidual=prename+'.residual' | |||
imsize=250 | |||
cell='2.0arcsec' | |||
minpb=0.2 | |||
restfreq='115.271201800GHz' | |||
outframe='LSRK' | |||
spw='0~1' | |||
width='5km/s' | |||
start='1400km/s' | |||
nchan=70 | |||
robust=0.5 | |||
phasecenter='J2000 12h22m54.9 +15d49m10' | |||
</source> | |||
Setup stopping criteria | |||
<source lang="python"> | |||
# In CASA | |||
stop=3. # multiplier for rms | |||
</source> | |||
Minimum size multiplier for beam area for removing very | |||
small mask regions. | |||
<source lang="python"> | |||
# In CASA | |||
pixelmin=0.5 # reasonable default is 1/2 the beam area | |||
</source> | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf '+prename+'.* ' +prename+'_*') | |||
clean(vis=vis,imagename=prename, | |||
imagermode='mosaic',ftmachine='mosaic',minpb=minpb, | |||
imsize=imsize,cell=cell,spw=spw, | |||
weighting='briggs',robust=robust,phasecenter=phasecenter, | |||
mode='velocity',width=width,start=start,nchan=nchan, | |||
restfreq=restfreq,outframe=outframe,veltype='radio', | |||
mask='', | |||
niter=0,interactive=F) | |||
</source> | |||
Inspect the resulting data cube: | |||
<source lang="python"> | |||
# In CASA | |||
viewer('M100.7m.CO.image') | |||
</source> | |||
Find properties of the dirty image | |||
<source lang="python"> | |||
# In CASA | |||
myimage=prename+'.image' | |||
bigstat=imstat(imagename=myimage) | |||
peak= bigstat['max'][0] | |||
print 'peak in cube = '+str(peak) | |||
thresh = peak /2. # sets loop threshold | |||
</source> | |||
== Determine or Set rms == | |||
In order to find properties of the dirty image, we will check the rms | |||
<source lang="python"> | |||
# In CASA | |||
if True: | |||
# If True: find the rms in two line-free channels | |||
chanstat=imstat(imagename=myimage,chans='4') | |||
rms1= chanstat['rms'][0] | |||
chanstat=imstat(imagename=myimage,chans='66') | |||
rms2= chanstat['rms'][0] | |||
rms=0.5*(rms1+rms2) | |||
else: | |||
# Set rms by hand | |||
rms=0.013 | |||
</source> | |||
== Make Final Images == | |||
<source lang="python"> | |||
# In CASA | |||
print 'rms in a channel = '+str(rms) | |||
</source> | |||
Deterimine the beam area in pixels for later removal of very small mask regions | |||
<source lang="python"> | |||
# In CASA | |||
major=imhead(imagename=myimage,mode='get',hdkey='beammajor')['value'] | |||
minor=imhead(imagename=myimage,mode='get',hdkey='beamminor')['value'] | |||
pixelsize=float(cell.split('arcsec')[0]) | |||
beamarea=(major*minor*pi/(4*log(2)))/(pixelsize**2) | |||
print 'beamarea in pixels =', beamarea | |||
n=-1 | |||
while (thresh >= stop*rms): | |||
n=n+1 | |||
print 'clean threshold this loop is', thresh | |||
threshmask = prename+'_threshmask' +str(n) | |||
maskim = prename+'_fullmask' +str(n) | |||
immath(imagename = [myresidual], | |||
outfile = threshmask, | |||
expr = 'iif(IM0 > '+str(thresh) +',1.0,0.0)', | |||
mask=myflux+'>'+str(minpb)) | |||
if (n==0): | |||
os.system('cp -r '+threshmask+' '+maskim+'.pb') | |||
print 'This is the first loop' | |||
else: | |||
makemask(mode='copy',inpimage=myimage, | |||
inpmask=[threshmask,mymask], | |||
output=maskim) | |||
imsubimage(imagename=maskim, mask=myflux+'>'+str(minpb), | |||
outfile=maskim+'.pb') | |||
print 'Combined mask ' +maskim+' generated.' | |||
os.system('cp -r '+maskim+'.pb ' +maskim+'.pb.min') | |||
maskfile=maskim+'.pb.min' | |||
ia.open(maskfile) | |||
mask=ia.getchunk() | |||
labeled,j=scipy.ndimage.label(mask) | |||
myhistogram = scipy.ndimage.measurements.histogram(labeled,0,j+1,j+1) | |||
object_slices = scipy.ndimage.find_objects(labeled) | |||
threshold=beamarea*pixelmin | |||
for i in range(j): | |||
if myhistogram[i+1]<threshold: | |||
mask[object_slices[i]] = 0 | |||
ia.putchunk(mask) | |||
ia.done() | |||
print 'Small masks removed and ' +maskim +'.pb.min generated.' | |||
os.system('rm -rf '+mymask+'') | |||
clean(vis=vis,imagename=prename, | |||
imagermode='mosaic',ftmachine='mosaic',minpb=minpb, | |||
imsize=imsize,cell=cell,spw=spw, | |||
weighting='briggs',robust=robust,phasecenter=phasecenter, | |||
mode='velocity',width=width,start=start,nchan=nchan, | |||
restfreq=restfreq,outframe=outframe,veltype='radio', | |||
mask = maskim+'.pb.min', | |||
interactive = F, | |||
niter = 10000, | |||
threshold = str(thresh) +'Jy/beam') | |||
if thresh==stop*rms: break | |||
thresh = thresh/2. | |||
# Run a final time with stop*rms if more than a little above stop*rms | |||
if (thresh < stop*rms and thresh*2.>1.05*stop*rms): | |||
thresh=stop*rms | |||
os.system('cp -r '+myimage+' '+myimage+str(n)) | |||
</source> | |||
== Image Analysis : Moment Maps == | |||
<figure id="M100.7m.CO1_0_Line.Clean.image.mom0.png"> | |||
[[Image:M100.7m.CO1_0_Line.Clean.image.mom0.png|200px|thumb|right|<caption>The CO(1-0) integrated intensity map (moment 0) of the M100 mosaic.</caption>]] | |||
</figure> | |||
<figure id="M100.7m.CO1_0_Line.Clean.image.mom1.png"> | |||
[[Image:M100.7m.CO1_0_Line.Clean.image.mom1.png|200px|thumb|right|<caption>The CO(1-0) velocity field (moment 1: color) and integrated intensity map (moment 0: contours) of the M100 mosaic.</caption>]] | |||
</figure> | |||
Next we will make moment maps for the CO(1-0) emission: Moment 0 is the integrated intensity; Moment 1 is the intensity weighted velocity field; and Moment 2 is the intensity weighted velocity dispersion. | |||
Above we determined the rms noise levels for M100 mosaics in a line-free and a line-bright channel. | |||
We want to limit the channel range of the moment calculations to those channels with significant emission. One good way to do this is to open the cube in the viewer overlaid with 3-sigma contours, with sigma corresponding to the line-free rms. | |||
<source lang="python"> | |||
# In CASA | |||
imview(raster={'file': 'M100.7m.CO1_0.image', | |||
'range': [-0.04,0.4]}, | |||
contour={'file': 'M100.7m.CO1_0.image', | |||
'levels': [0.004],'unit': 5}) | |||
</source> | |||
We find a channel range for significant emission of 10~61. | |||
For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity. | |||
<source lang="python"> | |||
# In CASA | |||
myimage='M100_7m_cube.image' | |||
chanstat=imstat(imagename=myimage,chans='4') | |||
rms1= chanstat['rms'][0] | |||
chanstat=imstat(imagename=myimage,chans='66') | |||
rms2= chanstat['rms'][0] | |||
rms=0.5*(rms1+rms2) | |||
print 'rms in a channel = '+str(rms) | |||
os.system('rm -rf M100_7m_cube.image.mom*') | |||
immoments(imagename = 'M100_7m_cube.image', | |||
moments = [0], | |||
axis = 'spectral', | |||
chans = '10~61', | |||
box='65,50,195,182', | |||
includepix = [rms*2,100], | |||
outfile = 'M100_7m_cube.image.mom0') | |||
</source> | </source> | ||
==Continue on to | For higher order moments it is very important to set a conservative flux threshold. Typically something like 6sigma, using sigma from a bright line channel works well. We do this with the '''mask''' parameter in the commands below. When making multiple moments, {{immoments}} appends the appropriate file name suffix to the value of '''outfile'''. | ||
<source lang="python"> | |||
# In CASA | |||
immoments(imagename = 'M100_7m_cube.image', | |||
moments = [1], | |||
axis = 'spectral', | |||
chans = '10~61', | |||
box='65,50,195,182', | |||
includepix = [rms*5.5,100], | |||
outfile = 'M100_7m_cube.image.mom1') | |||
</source> | |||
Next we can see these moment maps from these images using '''imview'''. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf M100_7m_cube.mom*.png') | |||
Im0 = 'M100_7m_cube.image.mom0' | |||
Im1 = 'M100_7m_cube.image.mom1' | |||
imview(raster = {'file': Im0, | |||
'colorwedge': T, | |||
'colormap': 'Rainbow 2', | |||
'scaling': -1.6, | |||
'range': [0.1,140], | |||
'beamellipse':T}, | |||
out = 'M100_7m_cube.mom0.png') | |||
imview(raster = {'file': Im1, | |||
'colorwedge': T, | |||
'colormap': 'Rainbow 2'}, | |||
contour = {'file': Im0, | |||
'levels': [5,15,45,135], | |||
'base': 0, | |||
'unit': 1.0}, | |||
out = 'M100_7m_cube.mom1.png') | |||
</source> | |||
== Export data as fits == | |||
If you want to analyze the data using another software package it is easy to convert from CASA format to FITS. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf M100_7m_*.fits') | |||
exportfits(imagename='M100_7m_cube.image', fitsimage='M100_7m_cube.image.fits') | |||
exportfits(imagename='M100_7m_cube.image.mom0', fitsimage='M100_7m_mom0.fits') | |||
exportfits(imagename='M100_12m_cube.flux', fitsimage='M100_12m_cube.flux.fits') | |||
</source> | |||
Although "FITS format" is supposed to be a standard, in fact most packages expect slightly different things from a FITS image. If you are having difficulty, try setting '''velocity=T''' and/or '''dropstokes=T'''. | |||
==Continue on to Combining Images with SD and 12m dataset== | |||
Now you can continue on to the [[M100_Band3_Combine_4.1]]. | Now you can continue on to the [[M100_Band3_Combine_4.1]]. | ||
{{Checked 4.1.0}} | {{Checked 4.1.0}} | ||
Latest revision as of 16:38, 25 November 2014
This page is currently under construction.
DO NOT USE IT.
To navigate the CASAguides pages, visit [http://casaguides.nrao.edu/ casaguides.nrao.edu ]
- This guide requires CASA 4.1.0 and assumes that you have downloaded M100_Band3_7m_UncalibratedData.tgz from M100_Band3#Obtaining_the_Data
- Details of the ALMA observations are provided at M100_Band3
- This portion of the guide covers calibration of the raw visibility data. To skip to the imaging portion of the guide, see: M100_Band3_Combine_4.1.
Overview
This part of the M100 Band 3 7m CASA guide will take you through the process of fully calibrating the visibility data. We will begin by flagging (marking as bad) data known to be useless before any inspection, for example data where one telescope blocks the line of sight of another. Then we will apply telescope-generated calibration tables to partially correct for atmospheric effects. After inspecting the data, we will flag some additional data that exhibit pathologies. Then we will use observations of the calibrators Titan, 3c273, and J1215+1654 to derive the phase and amplitude response of individual antennas as a function of time and frequency ("phase", "amplitude", and "bandpass" calibrations). We will apply these to the data and then extract the calibrated source data into a file appropriate for imaging.
The general procedure in this guide follows the other ALMA CASA guides: IRAS16293Band9, NGC3256Band3 and TWHydraBand7.
Unpack the Data
Once you have downloaded the M100_Band3_7m_UncalibratedData.tgz, unpack the file in a terminal outside CASA using
tar -xvzf M100_Band3_7m_UncalibratedData.tgz
then change directory to the new directory
cd M100_Band3_7m_UncalibratedData
You may wish to type
ls
to look at the files present. Note that a description of the observations can be found here: M100_Band3
The files are in native ALMA format (ALMA Science Data Model or ASDM) and will be converted to measurement sets using the CASA task importasdm below.
This guide requires Python module analysisUtils. If you have not already installed analysisUtils please follow the link to do so.
To begin, start CASA by typing
casapy
Be sure that you are using the correct CASA version, indicated at the top of this page.
Confirm your version of CASA
This guide has been written for CASA release 4.1.0. Please confirm your version before proceeding.
# In CASA
version = casadef.casa_version
print "You are using " + version
if (version < '4.1.0'):
print "YOUR VERSION OF CASA IS TOO OLD FOR THIS GUIDE."
print "PLEASE UPDATE IT BEFORE PROCEEDING."
else:
print "Your version of CASA is appropriate for this guide."
Install Analysis Utilities
Analysis Utilities (or analysisUtils for short) is a small set of Python scripts that provide a number of analysis and plotting utilities for ALMA data reduction. This guide uses a few of these utilities. They are very easy to install (just download and untar). See
http://casaguides.nrao.edu/index.php?title=Analysis_Utilities
for a full description and download instructions. Analysis Utilities are updated frequently so if its been a while since you installed it, its probably worth doing it again. If you are at an ALMA site or ARC, the analysis utilities are probably already installed and up to date.
A Note on Setting up to use multiple datasets
We will calibrate the six datasets individually and concatenate them into a single dataset at the end, before proceeding with the imaging part.
To do this, we start by defining and array called "basename" consisting of the uid names of the six data files. This will allow us to step through the calibration of the six datasets, where appropriate, using these 'for' loops. We will not be able to show every diagnostic plot but we give an example of each and the syntax to generate the rest. Also, please be aware that even on a very fast machine this whole process can take a while, as we are dealing with a lot of data.
You only need to define your list of files once per CASA session. Then "basename" will be a variable in the casapy shell. You can check if it exists by typing "print basename". In the interests of allowing you to easily exit and restart CASA and pick this guide up at any point we will redefine "basename" in each section of the guide. Feel free to skip this step if you've already defined it in your session. Remember that if you log out of CASA and want to continue with the calibration using these arrays, you will have to re-issue this command.
# In CASA
# Define an array containing the 'uid' names of all of our data sets
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print name
Create Measurement Sets
The raw data have been provided to you in the ASDM format. It is the native format of the data produced by the observatory.
Before we can proceed to the calibration, we will need to convert those data to the CASA MS format. This is done simply with the task importasdm.
#In CASA
for name in basename:
importasdm(asdm = name, asis='*')
Initial Inspection
We will take stock of what we have and obtain summary information about each dataset (targets observed, time range, spectral setup, and so on) by using the task listobs to output .txt format files. If you have not already done so, begin by reviewing the description of the observations here: M100_Band3.
#In CASA
# Loop over each element in the list and create summary file using listobs
for name in basename:
os.system('rm -rf '+name+'.ms.listobs.txt')
listobs(vis=name+'.ms', listfile=name+'.ms.listobs.txt', verbose=True)
The "for" loop executes for each element in basename, calling listobs and directing the output to a file called, e.g., "uid___A002_X5e971a_X124.ms.listobs.txt" for the first measurement set.
Note that after cutting and pasting a 'for' loop like this you often have to press return twice to execute. You may also want to take care to paste a line at a time if you are having trouble copy and pasting. Even better, you can use "cpaste" to paste blocks of code. To do so type "cpaste" at the CASA prompt, paste your commands, and then type "--" and hit return on the final (otherwise empty) line. This should look something like this:
CASA <8>: cpaste Pasting code; enter '--' alone on the line to stop. :basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3', 'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb'] : :for name in basename: : print name :-- uid___A002_X5e971a_X124 uid___A002_X5e971a_X2e7 uid___A002_X5e9ff1_X3f3 uid___A002_X5e9ff1_X5b3 uid___A002_X60b415_X44 uid___A002_X62f759_X4eb CASA <9>:
cpaste should be much more robust than copying-and-pasting directly into the shell but if you have trouble, just carefully paste one line at a time directly into CASA and hit return until the desired command executes.
You can browse through the listobs output as you would normally look at a text file (use emacs, vi, or another editor). You can also send the output to the terminal from inside of CASA. To do so type:
# In CASA
os.system('cat uid___A002_X5e971a_X124.ms.listobs.txt')
or
# In CASA
os.system('more uid___A002_X5e971a_X124.ms.listobs.txt')
CASA knows a few basic shell commands like 'cat', 'ls', and 'rm' but for more complex commands you will need to run them inside 'os.system("command")'. For more information see http://casa.nrao.edu/ .
Here is an example of the (abridged) output from listobs for the first dataset in the list, uid___A002_X5e971a_X124.ms. You would see this if you had specified verbose to be False in the listobs call:
================================================================================
MeasurementSet Name: /lustre/naasc/almauser/M100Band3/uid___A002_X5e971a_X124.ms MS Version 2
================================================================================
Observer: cvlahakis Project: uid://A002/X5d9e5c/X5d
Observation: ALMA
Data records: 940680 Total integration time = 5436.77 seconds
Observed from 17-Mar-2013/04:07:32.8 to 17-Mar-2013/05:38:09.6 (UTC)
ObservationID = 0 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows nUnflRows SpwIds Average Interval(s) ScanIntent
17-Mar-2013/04:07:32.3 - 04:09:35.4 1 0 J1229+0203 43200 43200.00 [0, 1, 2, 3, 4, 5, 6, 7] [1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] CALIBRATE_POINTING#ON_SOURCE
04:10:57.2 - 04:11:57.7 2 0 J1229+0203 45360 45360.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_SIDEBAND_RATIO#ON_SOURCE
04:12:48.5 - 04:14:00.0 3 0 J1229+0203 12960 12960.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHERE#ON_SOURCE
04:14:44.2 - 04:24:49.9 4 0 J1229+0203 118800 44880.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_BANDPASS#ON_SOURCE
04:25:53.4 - 04:27:56.4 5 1 J1445-1629 43200 43200.00 [0, 1, 2, 3, 4, 5, 6, 7] [1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] CALIBRATE_POINTING#ON_SOURCE
04:29:02.6 - 04:30:13.9 6 2 Titan 12960 12960.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHERE#ON_SOURCE
04:30:57.7 - 04:36:00.5 7 2 Titan 59400 22468.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_AMPLI#ON_SOURCE
04:37:10.1 - 04:39:13.2 8 3 J1215+1654 43200 43200.00 [0, 1, 2, 3, 4, 5, 6, 7] [1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] CALIBRATE_POINTING#ON_SOURCE
04:39:58.8 - 04:40:59.3 9 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
04:42:08.6 - 04:43:20.0 10 4 M100 12960 12960.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHERE#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 5 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 6 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 7 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 8 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 9 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 10 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 11 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 12 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 13 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 14 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 15 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 16 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:44:04.3 - 04:50:43.7 11 17 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:51:04.3 - 04:52:04.8 12 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 5 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 6 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 7 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 18 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 19 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 20 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 21 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 22 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 23 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 24 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 25 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 26 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 27 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:59:35.7 - 05:00:36.2 14 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
05:01:55.1 - 05:03:06.5 15 4 M100 12960 12960.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHERE#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 8 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 9 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 10 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 11 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 12 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 13 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 14 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 15 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 16 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 17 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 18 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 19 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 20 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:10:50.7 - 05:11:51.2 17 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 5 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 6 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 7 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 8 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 9 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 10 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 21 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 22 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 23 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 24 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 25 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 26 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 27 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:19:22.0 - 05:20:22.6 19 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 20 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 21 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 22 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 23 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 24 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 25 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 26 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:52:35.8 - 04:59:15.3 13 27 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
04:59:35.7 - 05:00:36.2 14 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
05:01:55.1 - 05:03:06.5 15 4 M100 12960 12960.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHERE#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 8 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 9 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 10 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 11 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 12 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 13 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 14 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 15 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 16 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 17 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 18 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 19 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:03:22.2 - 05:10:01.6 16 20 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:10:50.7 - 05:11:51.2 17 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 5 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 6 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 7 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 8 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 9 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 10 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 21 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 22 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 23 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 24 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 25 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 26 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:12:22.0 - 05:19:01.4 18 27 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:19:22.0 - 05:20:22.6 19 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
05:21:41.7 - 05:22:53.4 20 4 M100 12960 12960.00 [8, 9, 10, 11, 12, 13, 14, 15] [0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHERE#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 11 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 12 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 13 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 14 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 15 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 16 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 17 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 18 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 19 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 20 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 21 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 22 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:23:37.6 - 05:30:17.0 21 23 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:30:37.5 - 05:31:38.1 22 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 5 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 6 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 7 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 8 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 9 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 24 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 25 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 26 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:32:08.4 - 05:36:44.7 23 27 M100 5940 2244.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] OBSERVE_TARGET#ON_SOURCE
05:37:09.6 - 05:38:10.1 24 3 J1215+1654 11880 4488.00 [16, 17, 18, 19, 20, 21, 22, 23] [10.1, 1.01, 10.1, 1.01, 10.1, 1.01, 10.1, 1.01] CALIBRATE_PHASE#ON_SOURCE
(nRows = Total number of rows per scan)
Fields: 28
ID Code Name RA Decl Epoch SrcId nRows nUnflRows
0 none J1229+0203 12:29:06.699720 +02.03.08.59820 J2000 0 220320 146400.00
1 none J1445-1629 14:45:53.376290 -16.29.01.61880 J2000 1 43200 43200.00
2 none Titan 14:36:50.919019 -12.33.38.18202 J2000 1 72360 35428.00
3 none J1215+1654 12:15:03.979130 +16.54.37.95700 J2000 2 126360 74616.00
4 none M100 12:22:54.899040 +15.49.20.57160 J2000 2 38880 38880.00
5 none M100 12:22:57.009007 +15.49.13.60358 J2000 3 23760 8976.00
6 none M100 12:22:57.009041 +15.49.58.08112 J2000 3 23760 8976.00
7 none M100 12:22:54.339884 +15.47.22.41016 J2000 3 23760 8976.00
8 none M100 12:22:54.339918 +15.48.06.88770 J2000 3 23760 8976.00
9 none M100 12:22:54.339952 +15.48.51.36524 J2000 3 23760 8976.00
10 none M100 12:22:54.339986 +15.49.35.84278 J2000 3 17820 6732.00
11 none M100 12:22:54.340020 +15.50.20.32032 J2000 3 17820 6732.00
12 none M100 12:22:51.670863 +15.47.44.64936 J2000 3 17820 6732.00
13 none M100 12:22:51.670897 +15.48.29.12690 J2000 3 17820 6732.00
14 none M100 12:22:51.670931 +15.49.13.60444 J2000 3 17820 6732.00
15 none M100 12:22:51.670966 +15.49.58.08198 J2000 3 17820 6732.00
16 none M100 12:22:49.001808 +15.47.22.41102 J2000 3 17820 6732.00
17 none M100 12:22:49.001842 +15.48.06.88856 J2000 3 17820 6732.00
18 none M100 12:22:49.001876 +15.48.51.36610 J2000 3 17820 6732.00
19 none M100 12:22:49.001911 +15.49.35.84364 J2000 3 17820 6732.00
20 none M100 12:22:49.001945 +15.50.20.32118 J2000 3 17820 6732.00
21 none M100 12:22:59.677959 +15.47.22.40931 J2000 3 17820 6732.00
22 none M100 12:22:59.677993 +15.48.06.88685 J2000 3 17820 6732.00
23 none M100 12:22:59.678027 +15.48.51.36439 J2000 3 17820 6732.00
24 none M100 12:22:59.678061 +15.49.35.84193 J2000 3 17820 6732.00
25 none M100 12:22:59.678095 +15.50.20.31947 J2000 3 17820 6732.00
26 none M100 12:22:57.008938 +15.47.44.64850 J2000 3 17820 6732.00
27 none M100 12:22:57.008973 +15.48.29.12604 J2000 3 17820 6732.00
Spectral Windows: (24 unique spectral windows and 1 unique polarization setups)
SpwID Name #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) BBC Num Corrs
0 ALMA_RB_03#BB_1#SW-01#FULL_RES 124 TOPO 91955.512 -15625.000 1937500.0 1 XX YY
1 ALMA_RB_03#BB_1#SW-01#CH_AVG 1 TOPO 90986.762 1937500.000 1937500.0 1 XX YY
2 ALMA_RB_03#BB_2#SW-01#FULL_RES 124 TOPO 93893.012 -15625.000 1937500.0 2 XX YY
3 ALMA_RB_03#BB_2#SW-01#CH_AVG 1 TOPO 92924.262 1937500.000 1937500.0 2 XX YY
4 ALMA_RB_03#BB_3#SW-01#FULL_RES 124 TOPO 102033.637 15625.000 1937500.0 3 XX YY
5 ALMA_RB_03#BB_3#SW-01#CH_AVG 1 TOPO 102986.762 1937500.000 1937500.0 3 XX YY
6 ALMA_RB_03#BB_4#SW-01#FULL_RES 124 TOPO 104033.637 15625.000 1937500.0 4 XX YY
7 ALMA_RB_03#BB_4#SW-01#CH_AVG 1 TOPO 104986.762 1937500.000 1937500.0 4 XX YY
8 ALMA_RB_03#BB_1#SW-01#FULL_RES 128 TOPO 101942.187 -15625.000 2000000.0 1 XX YY
9 ALMA_RB_03#BB_1#SW-01#CH_AVG 1 TOPO 100926.562 1781250.000 1781250.0 1 XX YY
10 ALMA_RB_03#BB_2#SW-01#FULL_RES 128 TOPO 103757.337 -15625.000 2000000.0 2 XX YY
11 ALMA_RB_03#BB_2#SW-01#CH_AVG 1 TOPO 102741.712 1781250.000 1781250.0 2 XX YY
12 ALMA_RB_03#BB_3#SW-01#FULL_RES 128 TOPO 111814.962 15625.000 2000000.0 3 XX YY
13 ALMA_RB_03#BB_3#SW-01#CH_AVG 1 TOPO 112783.712 1781250.000 1781250.0 3 XX YY
14 ALMA_RB_03#BB_4#SW-01#FULL_RES 128 TOPO 113689.962 15625.000 2000000.0 4 XX YY
15 ALMA_RB_03#BB_4#SW-01#CH_AVG 1 TOPO 114658.712 1781250.000 1781250.0 4 XX YY
16 ALMA_RB_03#BB_1#SW-01#FULL_RES 4080 TOPO 101945.850 -488.281 1992187.5 1 XX YY
17 ALMA_RB_03#BB_1#SW-01#CH_AVG 1 TOPO 100949.756 1992187.500 1992187.5 1 XX YY
18 ALMA_RB_03#BB_2#SW-01#FULL_RES 4080 TOPO 103761.000 -488.281 1992187.5 2 XX YY
19 ALMA_RB_03#BB_2#SW-01#CH_AVG 1 TOPO 102764.906 1992187.500 1992187.5 2 XX YY
20 ALMA_RB_03#BB_3#SW-01#FULL_RES 4080 TOPO 111811.300 488.281 1992187.5 3 XX YY
21 ALMA_RB_03#BB_3#SW-01#CH_AVG 1 TOPO 112806.906 1992187.500 1992187.5 3 XX YY
22 ALMA_RB_03#BB_4#SW-01#FULL_RES 4080 TOPO 113686.300 488.281 1992187.5 4 XX YY
23 ALMA_RB_03#BB_4#SW-01#CH_AVG 1 TOPO 114681.906 1992187.500 1992187.5 4 XX YY
Sources: 80
ID Name SpwId RestFreq(MHz) SysVel(km/s)
0 J1229+0203 0 - -
0 J1229+0203 1 - -
0 J1229+0203 2 - -
0 J1229+0203 3 - -
0 J1229+0203 4 - -
0 J1229+0203 5 - -
0 J1229+0203 6 - -
0 J1229+0203 7 - -
0 J1229+0203 8 - -
0 J1229+0203 9 - -
0 J1229+0203 10 - -
0 J1229+0203 11 - -
0 J1229+0203 12 - -
0 J1229+0203 13 - -
0 J1229+0203 14 - -
0 J1229+0203 15 - -
0 J1229+0203 16 100950 0
0 J1229+0203 17 100950 0
0 J1229+0203 18 102794.1 0
0 J1229+0203 19 102794.1 0
0 J1229+0203 20 112794.1 0
0 J1229+0203 21 112794.1 0
0 J1229+0203 22 114669.1 0
0 J1229+0203 23 114669.1 0
1 J1445-1629 0 - -
1 J1445-1629 1 - -
1 J1445-1629 2 - -
1 J1445-1629 3 - -
1 J1445-1629 4 - -
1 J1445-1629 5 - -
1 J1445-1629 6 - -
1 J1445-1629 7 - -
1 Titan 8 - -
1 Titan 9 - -
1 Titan 10 - -
1 Titan 11 - -
1 Titan 12 - -
1 Titan 13 - -
1 Titan 14 - -
1 Titan 15 - -
1 Titan 16 100950 0
1 Titan 17 100950 0
1 Titan 18 102794.1 0
1 Titan 19 102794.1 0
1 Titan 20 112794.1 0
1 Titan 21 112794.1 0
1 Titan 22 114669.1 0
1 Titan 23 114669.1 0
2 J1215+1654 0 - -
2 J1215+1654 1 - -
2 J1215+1654 2 - -
2 J1215+1654 3 - -
2 J1215+1654 4 - -
2 J1215+1654 5 - -
2 J1215+1654 6 - -
2 J1215+1654 7 - -
2 J1215+1654 16 100950 0
2 J1215+1654 17 100950 0
2 J1215+1654 18 102794.1 0
2 J1215+1654 19 102794.1 0
2 J1215+1654 20 112794.1 0
2 J1215+1654 21 112794.1 0
2 J1215+1654 22 114669.1 0
2 J1215+1654 23 114669.1 0
2 M100 8 - -
2 M100 9 - -
2 M100 10 - -
2 M100 11 - -
2 M100 12 - -
2 M100 13 - -
2 M100 14 - -
2 M100 15 - -
3 M100 16 100950 0
3 M100 17 100950 0
3 M100 18 102794.1 0
3 M100 19 102794.1 0
3 M100 20 112794.1 0
3 M100 21 112794.1 0
3 M100 22 114669.1 0
3 M100 23 114669.1 0
Antennas: 9:
ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m)
East North Elevation x y z
0 CM01 N602 7.0 m -067.45.17.4 -22.53.22.3 8.8026 -527.8556 22.1988 2225080.352214 -5440132.953723 -2481524.785064
1 CM02 J502 7.0 m -067.45.17.7 -22.53.23.0 2.1079 -549.4459 22.1451 2225070.958100 -5440127.669506 -2481544.654450
2 CM03 J503 7.0 m -067.45.17.4 -22.53.23.2 9.2488 -555.0633 22.1293 2225076.734603 -5440122.930506 -2481549.823442
3 CM04 N605 7.0 m -067.45.17.4 -22.53.23.9 9.6883 -575.8319 22.0821 2225074.066737 -5440115.246896 -2481568.938246
4 CM05 J506 7.0 m -067.45.17.9 -22.53.23.2 -4.9539 -555.3433 22.1258 2225063.547041 -5440128.203265 -2481550.079981
5 CM06 N606 7.0 m -067.45.17.1 -22.53.23.6 19.1996 -566.5626 22.0993 2225084.240791 -5440114.998068 -2481560.405534
6 CM07 N601 7.0 m -067.45.17.0 -22.53.22.5 21.0601 -532.5792 22.2041 2225091.003357 -5440126.617491 -2481529.138855
7 CM09 N603 7.0 m -067.45.17.7 -22.53.22.3 -0.0719 -527.8532 22.2212 2225072.146648 -5440136.333195 -2481524.791579
8 CM12 J504 7.0 m -067.45.17.0 -22.53.23.0 22.2032 -550.2530 22.1451 2225089.438350 -5440119.771735 -2481545.398029
This output shows that four sources were observed in each data set: J1229+0203(3c273), Titan, J1215+1654, and M100.
- M100 is the science target. Note that the source corresponds to a number of individual fields (see the Field ID column). There are 23 pointings for the M100 Mosaic.
- Titan is observed once and is supposed to be used to set the absolute flux scale of the data (missing in dataset X4eb). However, fluxes are not determined due to bad quality, we will not use Titan as an amplitude calibrator.
- J1229+0203(3c273) plays two roles: it will serve as our bandpass calibrator, to characterize the frequency response of the antennas, and will be used to set the absolute flux scale of the data.
- J1215+1654 is "gain calibrator" to track changes in the phase and amplitude response of the telescopes over time. Observations of J1215+1654 are interleaved with observations of the M100 mosaic.
The output also shows that the data contain many spectral windows. Using the labeling scheme in the listobs above these are:
- spw 16,spw 18,spw 20, and spw 22 hold our science data. These are "Frequency Domain Mode" (FDM) data with small (0.49 MHz) channel width and wide total bandwidth. As a result these have a lot of channels (4080). spw 22 holds the upper sideband (USB) data and includes the CO(1-0) line. We will focus on these data. (datasets X3f3 and X5b3 only have two science spectral windows: spw 8 and spw 10)
- spw 8, spw 10, spw 12, and spw 14 hold lower a resolution processing ("Time Domain Mode", TDM) of the data from the same part of the spectrum (baseband). These data have only 128 channels across 2 GHz bandwidth and so have a much coarser channel spacing than the FDM data. These are used to create the Tsys calibration tables. (datasets X3f3 and X5b3 only have two atmcal spectral windows: spw 4 and spw 6)
The final column of the listobs output in the logger gives the scan intent. Later we will use this information to flag the pointing scans and the hot and ambient load calibration scans.
We'll now have a look at the configuration of the antennas used to take the data using the task plotants (<xr id="uid___A002_X5e971a_X124.plotants.png"/>).
<figure id="uid___A002_X5e971a_X124.plotants.png">
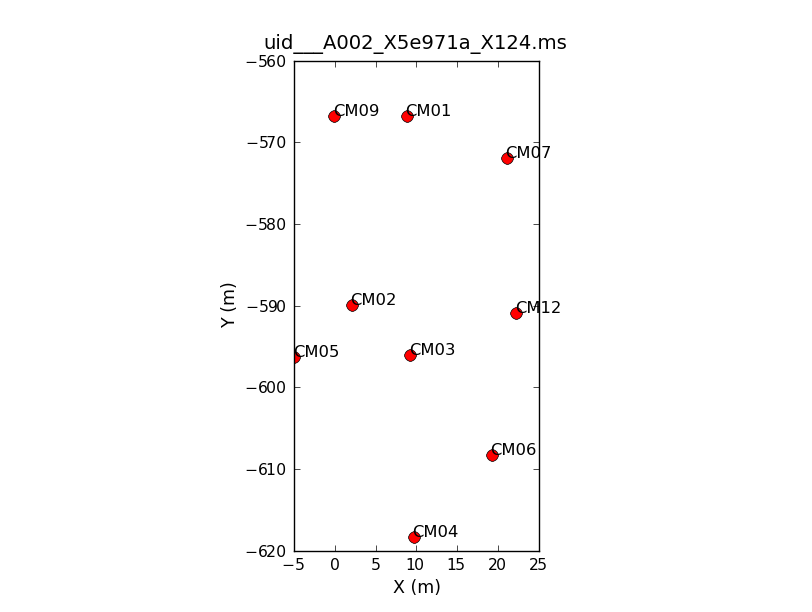
</figure>
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
os.system('rm -rf '+name+'.plotants.png')
print "Antenna configuration for : "+name
plotants(vis=name+'.ms', figfile=name+'.plotants.png')
dummy_string = raw_input("Hit <Enter> to see the antenna configuration for the next data set.")
This will loop through all 6 data sets, show you the antenna position for each, and save that as a file named, e.g., "uid___A002_X5e971a_X124.plotants.png" for the first data set. The "raw_input" command asks CASA to wait for your input before proceeding. If you would prefer to just browse the .png files after the fact you can remove this. Notice that the antenna setup changes, but only slightly, over the course of the 6 data sets.
A Priori Flagging
Even before we look in detail, we know that there are some data that we wish to exclude. We will start by flagging "shadowed" data where one antenna blocks the line of sight of another. We will also flag scans that were used to carry out pointing and atmospheric calibration, identified by their scan intent. Finally, we'll flag the autocorrelation data (the correlation of the signal from an antenna with itself) as we are only interested in cross-correlation data to make an interferometric image.
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
# In CASA
for name in basename:
print " "+name
flagcmd(vis=name+'.ms', inpmode='table', action='apply')
You may want to reset the flagging if you have tried this step before and are starting over though this is not necessary on your first time through. Do so using flagdata:
Then flag shadowed data using the command flagdata:
# In CASA
for name in basename:
print "Flagging shadowed data for "+asdm
flagdata(vis=name+'.ms',mode = 'shadow', flagbackup = F)
In the flagdata task we choose:
- vis = name+'.ms' : each measurement set
- mode = 'shadow': flag shadowed data
- flagbackup = F: Do not automatically back up the flag files. We will save all of the a priori flags together using flagmanager at the end of this subsection and save some space and time.
The relevant calibration information has already been extracted from the pointing and atmospheric scans and we will not need them below. Now flag the pointing scans using flagdata in 'manualflag' mode and selecting on 'intent':
# In CASA
for name in basename:
print "Flagging calibration scans for "+name
flagdata(vis=name+'.ms', mode='manual', intent='*POINTING*,*SIDEBAND_RATIO*,*ATMOSPHERE*', flagbackup = F)
Note that because the atmospheric calibration scans contain only TDM spectral windows, they will be removed automatically when we separate out the FDM data below.
Now flag the autocorrelation data:
# In CASA
for name in basename:
print "Flagging autocorrelation data for "+name
flagdata(vis=name+'.ms',autocorr=True,flagbackup=F)
Finally store the current flags information using flagmanager:
# In CASA
for name in basename:
print "Backing up 'a priori' flags for "+name
flagmanager(vis = name+'.ms', mode = 'save', versionname = 'Apriori')
The flagmanager task will also allow you to view the saved flagging versions, including those created by running flagdata with flagbackup=T. For example try flagmanager(vis='uid___A002_X5e971a_X124.ms', mode='list') The output in the logger should list the Apriori flagging that we have applied. It will also indicate versions associated with any flagdata command where you did not set flagbackup=F. Other tasks, for example applycal, will also create flag version tables.
Create and Apply Tsys and Antenna Position Calibration Tables
Tsys
The Tsys calibration gives a first-order correction for the atmospheric opacity as a function of time and frequency and associates weights with each visibility that persists through imaging.
Use gencal to create the Tsys calibration tables from the spectral windows with CALIBRATE_ATMOSPHERE intents in listobs. Later in the applycal stage this TDM Tsys table will be interpolated to the FDM.
#In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
os.system('rm -rf '+name+'.ms.tsys')
gencal(vis = name+'.ms',
caltable=name+'.ms.tsys',
caltype = 'tsys')
for i in range(0,6):
asdm = basename[i]
au.plotbandpass(caltable=asdm+'.ms.tsys', yaxis='amp', xaxis='freq',
buildpdf=False, figfile='./plots_tsys/tsysall'+spw+'_'+asdm+'.png', interactive=False,
antenna='CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12',chanrange='8~120',
subplot=42,overlay='antenna')
This sequence loops over all of our files and plots Tsys as a function of time for channel. In the call to plotcal: The Tsys values in CM01 and CM07 are too high. They will be flagged.
- subplot=42 parameter sets up a 4 x 2 panel grid.
- iteration tells plotcal to make a separate plot for each antenna.
The 'raw_input' commands will wait for you to hit Enter before issuing the next plot command. In the example above these are commented out (the leading "#" means that CASA will ignore them). If you would like to interactively cycle through the plots, uncomment them by removing the "#". Otherwise, the figfile parameter directs the output to .png files for later inspection. The easiest way to look at the 20 plots produced here is to simply inspect the .png files using your favorite viewer.
We will also want to look at Tsys as a function of frequency. This will use the analysisutils package mentioned at the beginning of this guide (called by the au. command)
<figure id="uid___A002_X5e971a_X124.spw8.t0.png">
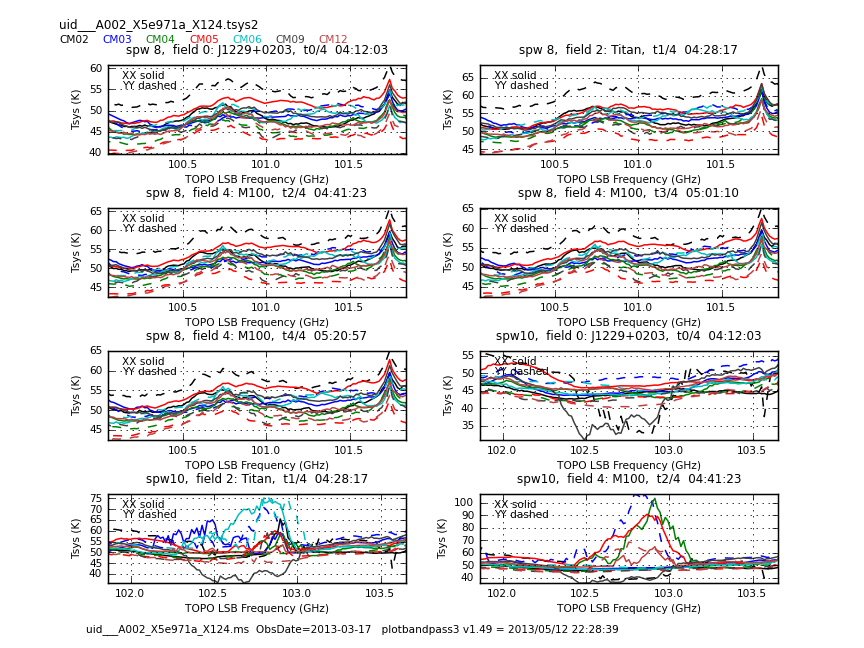
</figure> <figure id="uid___A002_X5e971a_X124.spw10.t3.png">
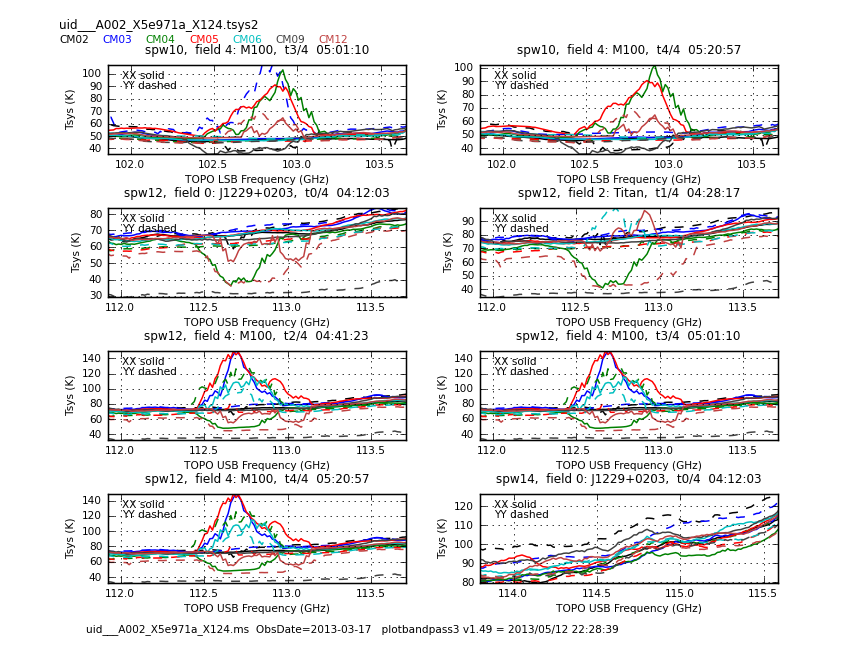
</figure>
<figure id="uid___A002_X5e971a_X124.spw14.t1.png">
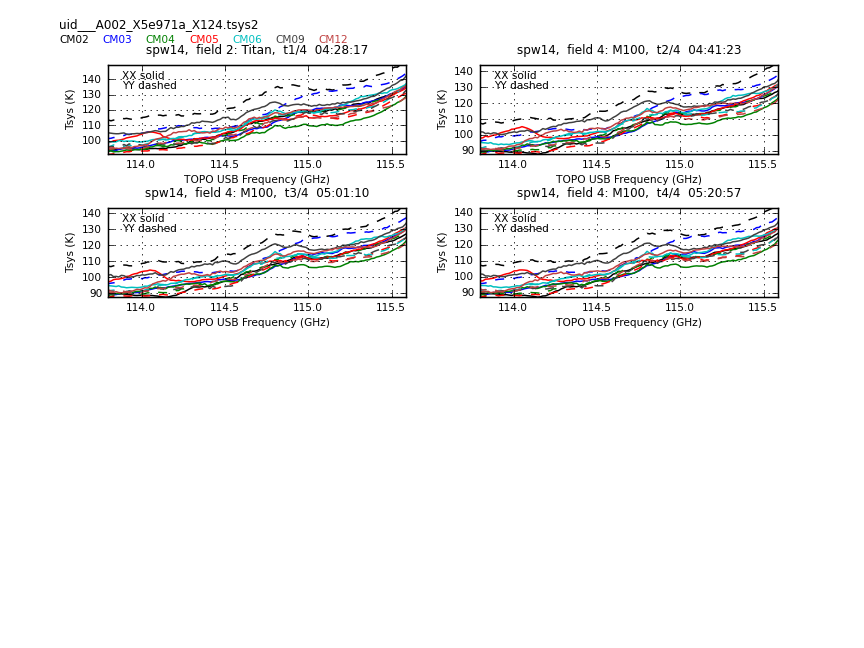
</figure>
Now have a look at the Tsys vs. frequency plots or see <xr id="uid___A002_X5e971a_X124.spw8.t0.png"/>, <xr id="uid___A002_X5e971a_X124.spw10.t3.png"/>, and <xr id="uid___A002_X5e971a_X124.spw14.t1.png"/> for examples on the first data set. You can see the effect of a close pair of atmospheric ozone absorption lines at about 101.8 GHz that makes Tsys larger near that frequency in all antennas. Applying the Tsys calibration tables will minimize the contribution of these atmospheric lines.
Additional issues: We will flag the spikes, which are appearing in Tsys caltable.
#In CASA
flagdata(vis = 'uid___A002_X60b415_X44.ms.tsys',
mode = 'manual',
antenna = 'CM09', spw = '8:32',
flagbackup = T)
flagdata(vis = 'uid___A002_X60b415_X44.ms.tsys',
mode = 'manual',
antenna = 'CM09', spw = '8:96',
flagbackup = T)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys',
mode = 'manual',
antenna = 'CM02', spw = '10:94~97',
flagbackup = T)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys',
mode = 'manual',
antenna = 'CM02', spw = '12:93~97',
flagbackup = T)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys',
mode = 'manual',
antenna = 'CM06', spw = '10:96~97',
flagbackup = T)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys',
mode = 'manual',
antenna = 'CM06', spw = '12:95~97',
flagbackup = T)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys',
mode = 'manual',
antenna = 'CM07', spw = '10:99~101',
flagbackup = T)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms.tsys',
mode = 'manual',
antenna = 'CM07', spw = '12:99~101',
flagbackup = T)
X124 and X2e7 have erratic behavior over parts of Tsys spectral window for spw=10 and 12: applying Tsys 'as is' produces artifacts, visible e.g. in amp vs chan plots for the phase calibrator
(which look reasonable before applying Tsys).
#In CASA
flagdata(vis = 'uid___A002_X5e971a_X124.ms.tsys',
mode = 'manual',
spw = '10:38~90',
flagbackup = T)
flagdata(vis = 'uid___A002_X5e971a_X124.ms.tsys',
mode = 'manual',
spw = '12:38~90',
flagbackup = T)
flagdata(vis = 'uid___A002_X5e971a_X2e7.ms.tsys',
mode = 'manual',
spw = '10:38~90',
flagbackup = T)
flagdata(vis = 'uid___A002_X5e971a_X2e7.ms.tsys',
mode = 'manual',
spw = '12:38~90',
flagbackup = T)
Antenna Positions
The antenna position table reflects refinements in the measured positions of the antennas from those stored in the data. gencal will now be used put antenna position data into each observation. Again, gencal will merely append to existing antenna position data, ruining any subsequent results. We start by removing any existing antenna position refinements, followed by defining the antenna names, then their refinements (both as arrays), finally running gencal to create the information CASA can refer to for antenna positions.
Note:Although four dataset (X124, X2e7, X3f3 and X5b3) have bad parameters of antenna positions, one dataset (X44) has correct parameters of antenna position, so we estimated "parameters" in gencal of four dataset using the antenna position table of X44 as a reference. X4eb has very good antenna position because the observation was done under good baseline condition (parameter in gencal is zero). Antenna position corrections are not necessary for X4eb.
#In CASA
os.system('rm -rf *.ms.antpos')
gencal(vis = 'uid___A002_X5e971a_X124.ms',
caltable = 'uid___A002_X5e971a_X124.ms.antpos',
caltype = 'antpos',
antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12',
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,
1.04838982e-05,4.57765535e-04,3.53057869e-04,
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,
-8.91945325e-04,4.27036546e-04,1.40665658e-03,
-1.16256997e-04,2.12460477e-03,-5.08893328e-03,
-3.65406508e-03,1.86459431e-02,5.67199755e-03,
8.04639747e-03,4.01791865e-02,1.46969082e-03,
-5.75850718e-05,6.55882061e-04,7.88089819e-05])
gencal(vis = 'uid___A002_X5e971a_X2e7.ms',
caltable = 'uid___A002_X5e971a_X2e7.ms.antpos',
caltype = 'antpos',
antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12',
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,
1.04838982e-05,4.57765535e-04,3.53057869e-04,
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,
-8.91945325e-04,4.27036546e-04,1.40665658e-03,
-1.16256997e-04,2.12460477e-03,-5.08893328e-03,
-3.65406508e-03,1.86459431e-02,5.67199755e-03,
8.04639747e-03,4.01791865e-02,1.46969082e-03,
-5.75850718e-05,6.55882061e-04,7.88089819e-05])
gencal(vis = 'uid___A002_X5e9ff1_X3f3.ms',
caltable = 'uid___A002_X5e9ff1_X3f3.ms.antpos',
caltype = 'antpos',
antenna = 'CM02,CM03,CM04,CM05,CM06,CM07,CM09,CM12',
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,
1.04838982e-05,4.57765535e-04,3.53057869e-04,
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,
-8.91945325e-04,4.27036546e-04,1.40665658e-03,
-1.16256997e-04,2.12460477e-03,-5.08893328e-03,
-3.65406508e-03,1.86459431e-02,5.67199755e-03,
8.04639747e-03,4.01791865e-02,1.46969082e-03,
-5.75850718e-05,6.55882061e-04,7.88089819e-05])
gencal(vis = 'uid___A002_X5e9ff1_X5b3.ms',
caltable = 'uid___A002_X5e9ff1_X5b3.ms.antpos',
caltype = 'antpos',
antenna = 'CM02,CM03,CM04,CM05,CM06,CM09,CM12',
parameter = [-1.04341656e-04,4.29447740e-04,4.12447378e-04,
1.04838982e-05,4.57765535e-04,3.53057869e-04,
1.38600077e-03,-6.59998506e-04,-4.26100381e-03,
-8.91945325e-04,4.27036546e-04,1.40665658e-03,
-1.16256997e-04,2.12460477e-03,-5.08893328e-03,
8.04639747e-03,4.01791865e-02,1.46969082e-03,
-5.75850718e-05,6.55882061e-04,7.88089819e-05])
gencal(vis = 'uid___A002_X60b415_X44.ms',
caltable = 'uid___A002_X60b415_X44.ms.antpos',
caltype = 'antpos',
antenna = 'CM12,CM02,CM03,CM05',
parameter = [5.70082944295e-05,-0.00065711393962,-7.91980095891e-05,0.000104837426825,
-0.000430660050663,-0.000411915081903,-1.1060689293e-05,-0.000458996548728,
-0.00035344706733,0.000982235185802,-0.000412690453231,-0.00145998690277])
gencal(vis = 'uid___A002_X62f759_X4eb.ms',
caltable = 'uid___A002_X62f759_X4eb.ms.antpos',
caltype = 'antpos',
antenna = 'CM09',
parameter = [0,0,0])
Applying antpos and Tsys tables
We are now ready to apply the Tsys tables to the data with applycal, which reads the specified gain calibration tables, applies them to the (raw) data column, and writes the calibrated results into the corrected column. Again, we loop through all the datasets. It is important to only apply Tsys obtained close in time to the data being corrected, so in addition to looping over data sets we define the list of unique source names and loop over these. Then by setting gainfield and field to the same value we ensure that Tsys are only applied to the source for which they are measured. The applycal task now has much more flexibility for interpolating and applying calibrations derived in one spectral window to another, even if they do not share the same spectral shape (number of channels and channel width). This new functionality is used below to interpolate the TDM (128 channel) Tsys measurements to the FDM (4080 channel) spectral windows. This is controlled through the spectral window mapping parameter *spwmap*. Because this can be a bit confusing, we've written a "helper" function that will tell you what you should put for the Tsys calibration table part of spwmap. We only need to run it on one of the datasets because they are all the same in this regard.
# Re-entering our array
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
Since some datasets have 4 spws and other datasets have 2 spws the application of the calibration tables needs to be treated split into several steps. Also, X5b3 has the Tsys for the science fields done on the phase calibrator (the other datasets have Tsys done on M100 itself) and X44 does not have an observation of Titan as its amplitude calibrator, so we also need to treat these separately.
Now run the applycal commands.
#In CASA
from recipes.almahelpers import tsysspwmap
# When you run tsysspwmap(vis=name+'.ms',tsystable=name+'.ms.tsys') this will print, for example for uid___A002_X5e971a_X124:
# Datasets with 4 spws
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1229+0203',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
interp = 'linear, linear',
gainfield=['J1229+0203',''],
calwt=T,
flagbackup=F)
# Datasets where the Tsys is done on M100
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X62f759_X4eb']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1215+1654,M100',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
interp = 'linear, linear',
gainfield=['M100'],
calwt=T,
flagbackup=F)
# Dataset where Tsys is done on the phase calibrator rather than M100
for name in ['uid___A002_X60b415_X44']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1215+1654,M100',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
interp = 'linear, linear',
gainfield=['J1215+1654'],
calwt=T,
flagbackup=F)
# Datasets that have observations of Titan
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'Titan',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='Titan',
interp='linear,linear',
calwt=T,
flagbackup=F)
# Datasets with 2 spws
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1229+0203',
spw='8,10',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='J1229+0203',
interp='linear,linear',
calwt=T,
flagbackup=F)
applycal(vis=name+'.ms', field = 'Titan',
spw='8,10',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='Titan',
interp='linear,linear',
calwt=T,
flagbackup=F)
applycal(vis=name+'.ms', field = 'J1215+1654,M100',
spw='8,10',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='M100',
interp='linear,linear',
calwt=T,
flagbackup=F)
where:
- field: the field to which we will apply the calibration,
- gainfield: the field from which we wish to take the calibration table
Inspect Data
We are not quite done with the original ".ms" data sets yet. Before going further it will be useful to use plotms to show the effects of applying the calibration. In the process we'll take a quick look at each antenna and search for pathologies in the data.
For this basic inspection, we want to compare the phase and amplitude as a function of frequency and time in the DATA and CORRECTED columns of each measurement set. The CORRECTED column has had the Tsys applied and so we expect lower phase scatter and flatter amplitude response as a function of time and frequency. We are looking for antenna-based issues, so cycling through a set of baselines that includes each antenna once will be a good start. We'll focus these plots on the bandpass calibrator, 3c273, and on baselines that include antenna CM04, which we will make our reference antenna in just a bit.
Each CASA Measurement Set has up to three "columns" of data: DATA, CORRECTED, and MODEL (though it is possible for a MS to hold only a DATA column if it has not been processed at all). A column entry exists for each data point (baseline, frequency, time). The DATA column holds the current raw data, though using split as we just did we can change the definition of "raw" data. The CORRECTED column holds the result of applying one or more calibration tables (e.g., via applycal) to the DATA column and so represents a corrected version of the DATA column. In CASA 4.1: the MODEL column is deprecated (though it is still possible to use them by explicitly setting usescratch=T). Instead, the model is stored in the header of the ms. This is a great step forward in terms of saving space and time to create scratch columns To get an intuitive feel for the columns you may wish to explore using plotms (which can plot different data columns, as seen below) or the browsetable task.
<figure id="plotms_amp_vs_freq.png">
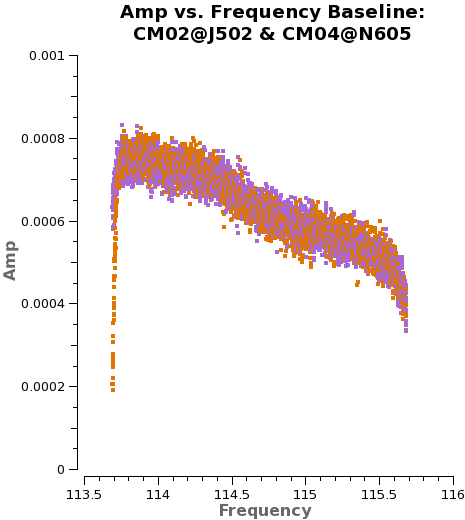
</figure> <figure id="plotms_amp_vs_freq_corr.png">
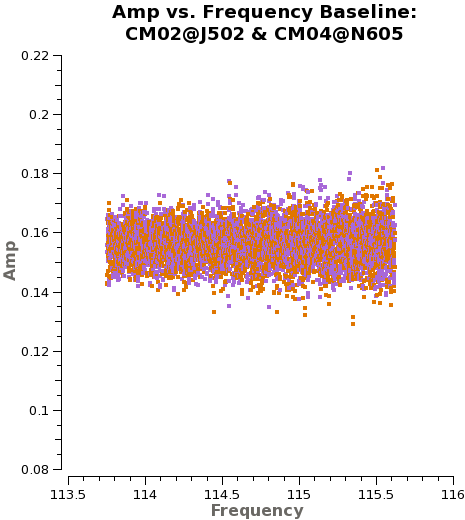
</figure>
First, we plot amplitude as a function of frequency for 3c273. We start by plotting the DATA column, set color to indicate the two correlations (i.e., the XX and YY polarizations), and ask plotms to iterate over baseline. By setting antenna to 'CM04&*' we select only baselines that include CM04. We ask plotms to average all data over a very long timescale, avgtime = 1e8 seconds. By setting avgscan = True we allow plotms to average across scan boundaries. The result is a plot of average amplitude per channel vs. frequency.
Here, we will show the dataset with 4BBs(X124, X2e7, X44, and X4eb) and 2BBs(X3f3 and X5b3), separately. For 4BBs dataset,
# In CASA
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
plotms(vis=name+'.ms',
field='J1229+0203',
xaxis='frequency', yaxis='amp',
selectdata=T, spw='22',
avgtime='1e8',avgscan=T,
coloraxis='corr',
iteraxis='baseline',
antenna='CM04&*',
ydatacolumn='data')
The rest dataset are here.
# In CASA
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
plotms(vis=name+'.ms',
field='J1229+0203',
xaxis='frequency', yaxis='amp',
selectdata=T, spw='10',
avgtime='1e8',avgscan=T,
coloraxis='corr',
iteraxis='baseline',
antenna='CM04&*',
ydatacolumn='data')
Notice the green arrows along the bottom of the plotms window. We asked plotms to iterate over baseline. As you click the arrows, the plot will rotate from baseline to baseline, always with CM04 so that each antenna shows up once. To see the effect of the calibration, go to the "Axes" tab along the left of the plotms window and pull down the Data Column menu under the Y Axis. Set this from DATA to CORRECTED and you should see the effects of the calibration. You may need to ensure that the "Force Reload" box is checked before clicking "Plot" (both buttons lie at the bottom of the panel). For the most part things get better (flatter).
<figure id="plotms_amp_vs_time.png">
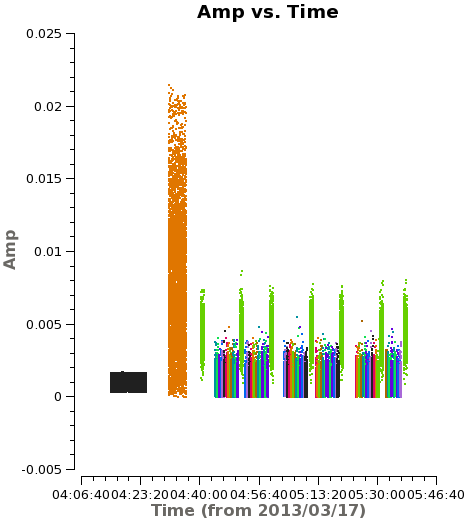
</figure> <figure id="plotms_amp_vs_time_corr.png">
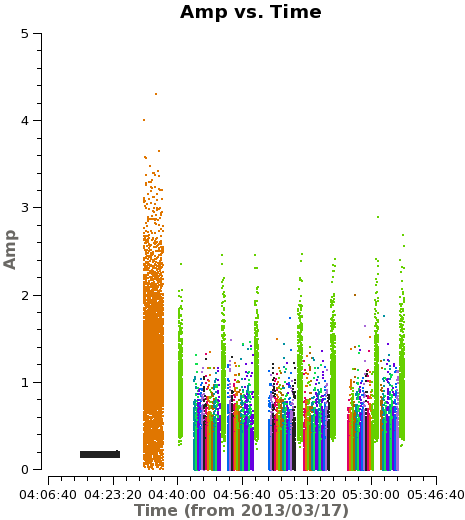
</figure>
You can now make analogous calls to examine the phase vs. frequency, amplitude vs. time, and phase vs. time. Here, we will show the examples of 4BBs dataset (X124 for this case).
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
name=basename[0]
plotms(vis=name+'.ms',
field='J1229+0203',
xaxis='frequency', yaxis='phase',
selectdata=T, spw='22',
avgtime='1e8',avgscan=T,
coloraxis='corr',
iteraxis='baseline',
antenna='CM04&*',
ydatacolumn='data')
plotms(vis=name+'.ms',
field='',
xaxis='time', yaxis='amp',
selectdata=T, spw='22:300~3800',
avgchannel='4000',avgscan=F,
coloraxis='field',
iteraxis='',
antenna='CM04&*',
ydatacolumn='data')
plotms(vis=name+'.ms',
field='',
xaxis='time', yaxis='phase',
selectdata=T, spw='22:300~3800',
avgchannel='4000',avgscan=F,
coloraxis='corr',
iteraxis='baseline',
antenna='CM04&*',
ydatacolumn='data')
Where:
- avgchannel set to a large number causes the plots of phase and amplitude vs. time to average data at all frequencies into a single point for each measurement (we exclude the channels at the edge of the spw as for those the Tsys is poorly measured).
- coloraxis corr sets the colors to correspond to the two polarizations of the data.
In each case, you will want to examine each baseline, alternating between the DATA and CORRECTED columns.
This is a lot of data inspection and that's only for one of 6 data sets! You can iterate across the data by hand, updating "asdm" to refer to each data set in order and cycling between baselines and DATA/CORRECTED. It is also possible to script CASA to show you the key plots in succession (see the next block down). However you approach the infrastructure, you are looking for:
- Improved scatter and lower variability in phase and amplitude vs. frequency and time. This indicates that Tsys (-> amp) and antenna position (-> phase) calibrations helped.
- Sudden jumps in phase or amplitude as a function of either time or frequency. These may indicate problems with the antenna during that observation.
- Large gradients, especially full wraps, in phase as a function of frequency. This may indicate a problem in the delays, the signal path length to the telescopes.
- Unusual magnitude, scatter, or patterns in any plot - though this may be better explored using plots that show all data together, which we'll make in a moment.
- Missing data. For example, if the phase calibrator drops out for a period of time we will not be able to calibrate and will need to flag the data.
As you look through, note individual potentially problematic antennas. If all antennas in a data set appear problematic it may be that your "reference" antenna, CM04 in the example above, is the source of the problem. In this case swap this reference antenna for another and see whether the problem is isolated to your original reference antenna.
A brief aside on structure: This section (Data Inspection) and the next (Apply Flags) are closely linked. We will present them as two separate steps. The idea here is that you look through your data, note problems, and then write commands to flag problematic data. This mimics one approach to writing data reduction scripts for CASA, where you will group all flagging together into one convenient place. Other CASA guides take a different approach, interleaving flagging and plotting. There is no "right" answer here. Find an approach to data inspection that works for you.
If you do wish to semi-automate the plot generation, the following sequence will cycle between data and corrected plots for each data set in turn. Type "stop" at any input call to break out.
Note: In loops like this involving plotms, hitting enter before the last dataset completes loading may cause plotms to stop refreshing the plot window at each step, or cause the window to disappear altogether. This will be fixed in an upcoming CASA release. Right now, you may need to exit CASA and restart to bring up the plotms window again, but you should be able to continue in the script where you left off. Here, we will show the examples of 4BBs dataset.
# In CASA
user_input = ""
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
# check if a stop has been requested
if user_input == "stop":
break
# Extract antenna list for this data set.
tb.open(name+'.ms/ANTENNA', nomodify=True)
ants = tb.getcol('NAME')
tb.close
# Define the reference antenna to make baselines with
ref_ant = 'CM04'
# Loop over antennas
for ant in ants:
# Check if the user wants to stop
if user_input == "stop":
break
# Skip correlation of reference antenna with itself (autocorrelations are flagged anyhow)
if ant == ref_ant:
continue
# Define the baseline with the reference antenna for current antenna
ant_str = ref_ant+'&'+ant
print "Showing baseline "+ant_str+" for data set "+name
print "Use this to inspect effect of applying Tsys calibrations."
# Loop over phase and amplitude as axes
for y_axis in ["amp", "phase"]:
# Make 'before' plot for frequency x-axis
print "... "+y_axis+" vs. frequency for DATA:"
plotms(vis=name+'.ms', spw='22', field='J1229+0203',,
antenna=ant_str, xaxis="frequency", yaxis=y_axis,
avgtime="1e8", avgscan=T, coloraxis="corr",
ydatacolumn="data")
user_input = raw_input("Hit <ENTER> to see CORRECTED data [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
# Make 'after' plot for frequency x-axis
print "... "+y_axis+" vs. frequency for CORRECTED:"
plotms(vis=name+'.ms', spw='22', field='J1229+0203',,
antenna=ant_str, xaxis="frequency", yaxis=y_axis,
avgtime="1e8", avgscan=T, coloraxis="corr",
ydatacolumn="corrected")
user_input = raw_input("Hit <ENTER> to proceed to next plot [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
# Make 'before' plot for time x-axis
print "... "+y_axis+" vs. time for DATA:"
plotms(vis=name+'.ms', spw='22', field='J1229+0203',,
antenna=ant_str, xaxis="time", yaxis=y_axis,
avgchannel="1000", coloraxis="corr",
ydatacolumn="data")
user_input = raw_input("Hit <ENTER> to see CORRECTED data [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
# Make 'after' plot for time y-axis
print "... "+y_axis+" vs. time for CORRECTED:"
plotms(vis=name+'.ms', spw='22', field='J1229+0203',,
antenna=ant_str, xaxis="time", yaxis=y_axis,
avgchannel="1000", coloraxis="corr",
ydatacolumn="corrected")
user_input = raw_input("Hit <ENTER> to proceed to next plot [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
<figure id="plotms_amp_freq_M100.png">
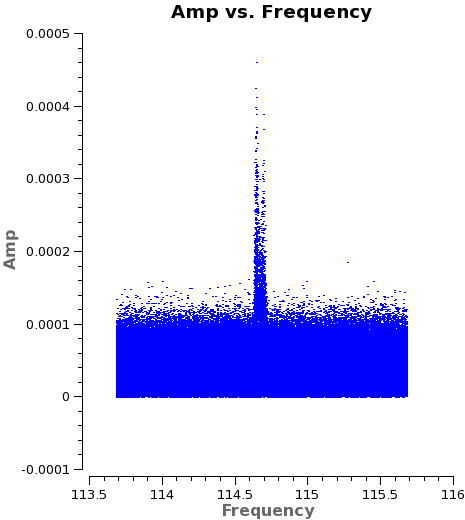
</figure>
A detailed explanation of the procedure is a bit outside the scope of this guide (for more on python see http://www.python.org/ and for more on the CASA toolkit see http://casa.nrao.edu/), but the basic process is to loop over each data set, baseline with the reference antenna (here CM04), and y-axis of interest (phase or amplitude) then plot the effect of the calibration vs. frequency and time for each combination. Running this to step through the data will give you about 200 "before and after" plots from which you could note a subset of problematic cases to be followed up by hand. Many other strategies to inspect the data are also viable.
With the Tsys applied successfully and the a priori flagging taken care of we will now split out the corrected data. We will keep only the corrected data, specified via datacolumn, and only spectral window 1, which contains the FDM (high spectral resolution) observations of the CO(1-0) line. Setting keepflags=F tells split not to carry over any fully flagged rows from the original data set to the new MS.
Next we will do a bit more inspection using plotms to look at whole data sets. This will help us identify missing data or look for egregious outliers.
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
plotms(vis=name+'.ms',
xaxis='time', yaxis='amp',
avgchannel='4080',coloraxis='field')
dummy_string = raw_input("Examining amplitude vs. time for "+name+" . Hit <Enter> to proceed.")
Here look for:
- Missing data. The source needs to be flanked by phase calibrator scans, if those are missing for any reason we need to flag the appropriate time range.
- Dramatic outliers. Does the source suddenly get very bright or the otherwise bright calibrator appear anomalously faint for a brief time? This likely indicates problematic data that should be identified and flagged. You can use the "select" (box with green plus along the bottom row in plotms) and "locate" (magnifying glass) buttons in plotms to isolate and identify problem data (it will print to the log).
- Smooth variation with time. A sudden jump may indicate a problem and often the safest approach is to flag data near a discontinuity.
Look through the amplitudes vs. time for each data set (remember that we've already examined the phases vs. time and amplitude vs. time for individual baselines above). Titan is missing in X4eb dataset, we will skip this dataset.
# In CASA
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44']:
plotms(vis=name+'.ms',
field='J1229+0203',
xaxis='uvdist', yaxis='amp',
avgchannel='4080',coloraxis='corr')
dummy_string = raw_input("Examining amplitude vs. UV Distance for J1229+0203 for "+name+" . Hit <Enter> to proceed.")
plotms(vis=name+'.ms',
field='Titan',
xaxis='uvdist', yaxis='amp',
avgchannel='4080',coloraxis='corr')
dummy_string = raw_input("Examining amplitude vs. UV Distance for Titan for "+name+" . Hit <Enter> to proceed [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
For this command notice that we can see the CO(1-0) line in the M100 even before calibration (see <xr id="plotms_amp_freq_M100.png"/>) shows evidence of a strong line. This suite of plots (along with the earlier inspection of the Tsys tables) gives us the tools we need to identify problematic data through the data sets. We use this to generate a set of inspection-driven flagdata commands for each data set. We apply these before the bandpass and gain calibration.
Apply Flagging
Based on this inspection and the other plots we have made, we now flag problematic portions of the data. We break up the flags by reason for flagging in order to illustrate the process. As you reduce your own data it may be more efficient to group flags by data set and make use of the flagcmd command. Except for the "post-calibration" flagging, the inspection plots that we just looked through have already revealed all of the problems that we flag. We structure the guide so that the flagging is grouped in one place.
As before, we may wish to reset our flags before beginning (particularly if one iterates this process) via:
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
flagdata(vis = name+'.ms',mode='unflag', flagbackup = F)
Remember that we dropped the flagged data when splitting out NO SPLIT SO FAR after the Tsys calibration, so this should not undo your "A Priori" flagging of shadowed data, autocorrelations, etc.. In any case this unflagging step is not necessary during the first iteration.
- Edge Channels
ALMA's sensitivity decreases near the edge of the baseband and it is often useful to check for a 'roll-off' in sensitivity near the edge of the band. This will appear as a downturn in amplitude as a function of channel near the edge of the window in the uncalibrated data, as a flaring due to increased noise at the spw edges in the calibrated data.
<figure id="plotms_CM04&CM06_edge.png">
</figure>
# In CASA
for name in basename:
flagdata(vis = name+'.ms', mode='manual', spw = '*:0~127,*:3952~4079', flagbackup = F)
- Problematic Tsys measurements
Above we noted issues with the Tsys measurements for both CM01 and CM07. CM07 only for 2 datasets? see below We flag the affected data.
# In CASA
for name in basename:
flagdata(vis= name+'.ms', mode='manual', antenna='CM01', flagbackup=F)
for name in basename:
flagdata(vis= name+'.ms', mode='manual', antenna='CM07', flagbackup=F)
Flagging of individual dataset: X4eb : there are spikes in spw=18 and 20 for CM02,CM06,CM07.
# In CASA
flagdata(vis = 'uid___A002_X62f759_X4eb.ms',
mode = 'manual',
antenna = 'CM02',
spw = '18:3080~3110',
flagbackup=F)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms',
mode = 'manual',
antenna = 'CM02',
spw = '20:3010~3105',
flagbackup=F)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms',
mode = 'manual',
antenna = 'CM06',
spw = '18:3080~3128',
flagbackup=F)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms',
mode = 'manual',
field = 'J1229+0203',
antenna = 'CM06',
spw = '20:3020~3120',
flagbackup=F)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms',
mode = 'manual',
field = 'J1229+0203',
antenna = 'CM07',
spw = '18:3160~3250',
flagbackup=F)
flagdata(vis = 'uid___A002_X62f759_X4eb.ms',
mode = 'manual',
field = 'J1229+0203',
antenna = 'CM07',
spw = '20:3140~3240',
flagbackup=F)
Flagging of individual dataset:
X3f3 & X5b3 have low amplitude of CM07
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for i in [2,3]:
name = basename[i]
print 'Flagging antenna CM07 on ms '+str(i)+': '+asdm+'.ms'
flagdata(vis = name+'.ms',
mode = 'manual',
antenna = 'CM07',
flagbackup=F)
- Unreliable Short-Spacing Measurements on Titan
<figure id="plotms_amp_uvdist_titan.png">
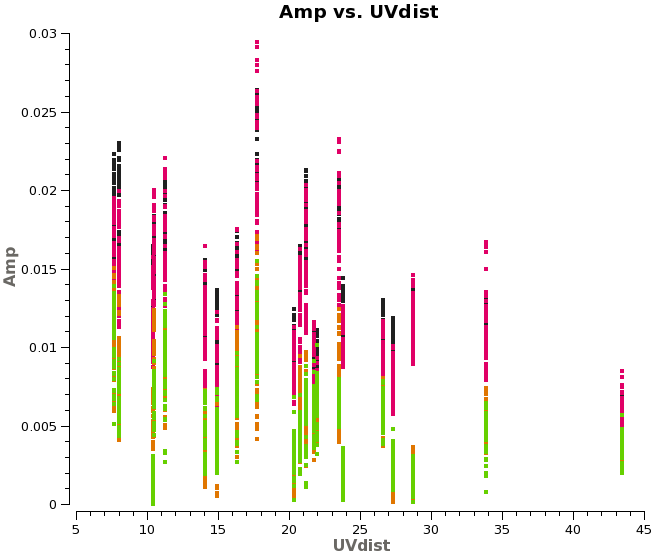
</figure> Saturn may contaminate the short u-v spacings from Titan. In any case these often show significant scatter (<xr id="plotms_amp_uvdist_titan.png"/>), so we flag them.
# In CASA
for name in basename:
flagdata(vis=name+'.ms', mode='manual', field='Titan', flagbackup = F)
Now that we've applied our flagging, back up the flags as version "User" using flagmanager:
# In CASA
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44']:
flagmanager(vis=name+'.ms',mode='save',versionname ='User')
Applying this flagging will remove the most egregious pathologies from the data. We are now ready to calibrate the data.
Bandpass Calibration
We begin by calibrating the phase and amplitude response of each antenna as a function of frequency, called "bandpass calibration." We have already seen that the data contain smooth but systematic variations in both phase and amplitude as a function of frequency. We can see this again in a more compact form by plotting phase as a function of frequency for all baselines associated with each antenna (<xr id="plotms_phase_freq_3c273.png"/>).
<figure id="plotms_phase_freq_3c273.png">
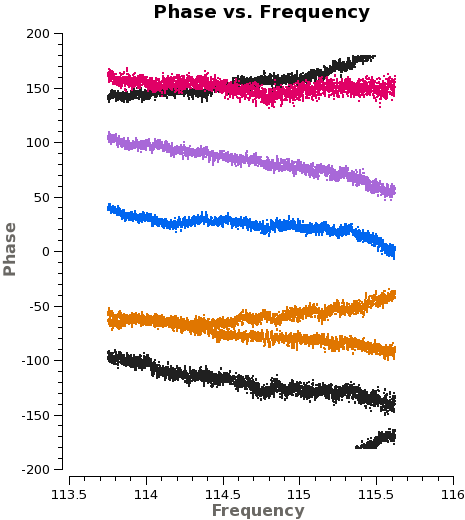
</figure>
# In CASA
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
plotms(vis= name+'.ms',
xaxis='freq', yaxis='phase', spw='22',
field='J1229+0203', correlation='XX',
avgtime='1e8', avgscan=T, antenna='*&*',
coloraxis='baseline')
dummy_string = raw_input("Plotting phase vs. frequency for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
# In CASA
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
plotms(vis= name+'.ms',
xaxis='freq', yaxis='phase', spw='10',
field='J1229+0203', correlation='XX',
avgtime='1e8', avgscan=T, antenna='*&*',
coloraxis='baseline')
dummy_string = raw_input("Plotting phase vs. frequency for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
Each plot shows phase as a function of frequency for all baselines with one antenna for 3c273(J1229+0203). We plot only the 'XX' correlation, colorizing by baseline. By using avgscan and a large avgtime we average all scans and integrations.
The phase (and amplitude) also varies as a function of time, as we saw before. Here are the similar plots for phase vs. time (see <xr id="plotms_phase_time_3c273.png"/>).
<figure id="plotms_phase_time_3c273.png">
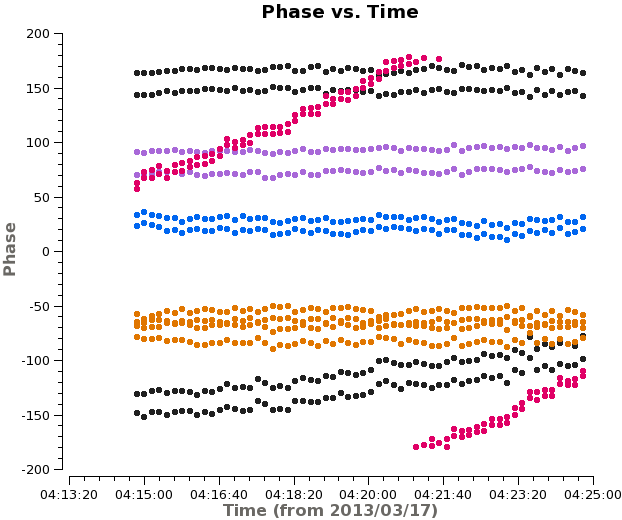
</figure>
# In CASA
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
plotms(vis= name+'.ms',
xaxis='time', yaxis='phase',
selectdata=True, field='J1229+0203',
spw='22', antenna='*&*',correlation='XX',
avgchannel='1000',
coloraxis='baseline')
dummy_string = raw_input("Plotting phase vs. time for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
# In CASA
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
plotms(vis= name+'.ms',
xaxis='time', yaxis='phase',
selectdata=True, field='J1229+0203',
spw='10', antenna='*&*',correlation='XX',
avgchannel='1000',
coloraxis='baseline')
dummy_string = raw_input("Plotting phase vs. time for "+name+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
<xr id="plotms_phase_time_3c273.png"/> shows that the phase varies with time. We need to take this temporal variation into account when we solve for the frequency variations. Therefore we carry out the bandpass calibration in two steps. First, we use gaincal to solve for the variation of phase as a function of time for 3c273(J1229+0203) on very short timescales. We set gaincal to derive a separate phase solution for each antenna every integration by setting solint to 'int'. We solve, averaging together only a small fraction of the total bandpass to avoid the effects of the phase vs. frequency behavior. We will then apply this solution to remove time-dependent behavior when we solve for the frequency response of the antennas with bandpass.
An integration is the smallest time quantum of observation in the data, usually a few seconds. You can check the integration length for each scan in listobs.
#In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print "Running a short solution interval phase calibration for "+name
os.system('rm -rf '+name+'.bpphase.gcal')
gaincal(vis = name+'.ms',
selectdata=T,field = 'J1229+0203',spw = '',
caltable = name+'.bpphase.gcal',
solint = '30.3s',refant = 'CM04', minsnr = 2.0, minblperant = 3)
Now we use bandpass to solve for the frequency response of each antenna. To do this, we average all data in time by setting solint to 'inf' (that is, 'infinite'). We allow combination across scans and the different field IDs found for 3c273 by setting combine to "scan,field". We apply the phase vs. time calibration that we just derived on-the-fly using the parameter "gaintable".
for name in basename:
print "Running a bandpass calibration for "+name
os.system('rm -rf '+name+'.bandpass.bcal')
bandpass(vis = name+'.ms',
field = 'J1229+0203',
gaintable = name+'.bpphase.gcal',
caltable = name+'.bandpass.bcal',
bandtype='B',
solint = 'inf, 16ch',combine = 'scan,field', solnorm=T,refant = 'CM04',
minsnr=1.0, minblperant = 2)
in these calls:
- caltable specifies the output calibration table
- gaintable specifies any calibration tables to be applied "on the fly" to the data before solving.
- solint sets the time interval for which solutions are derived for each antenna.
- refant set the reference antenna, here CM04, which is defined to have zero correction by construction.
- calmode 'p' sets gaincal to calibrate the phase only, other options are 'a'mplitude or 'ap' - amplitude and phase.
- minblperant sets the minimum baselines required per antenna for a successful solution.
- minsnr sets the minimum signal-to-noise ratio required for a successful solution.
- bandtype here 'B' tells bandpass to solve for the phase and frequency response of each antenna in each channel, averaging data in time across the solution interval.
- fillgaps tells bandpass to interpolate across channel gaps 1 channel wide.
- solnorm tells bandpass to normalize the bandpass amplitude and phase corrections to have magnitude unity. The absolute scale of the calibration will come from our later gaincal solutions.
Do not worry about the message "Insufficient unflagged antennas" when running the bandpass task. This indicates that bandpass is failing on the flagged edge channels, which is expected.
It is now a good idea to plot both sets of solutions to look for irregularities, especially:
- discontinuities in the phase vs. time solution
- rapid wrapping of phase in either phase vs. time or bandpass solution
- large roll-off in the amplitude response near the band edge in the bandpass solution
- large scatter in any solution.
We loop through and using analysis utils, again generating .png files of each calibration. As the bandpass plots take a while you may want to go have a cup of coffee and inspect them using your favorite image viewer. Otherwise, switch the interactive parameter to True to see them in real time instead.
<figure id="bandpass.CM02.png">
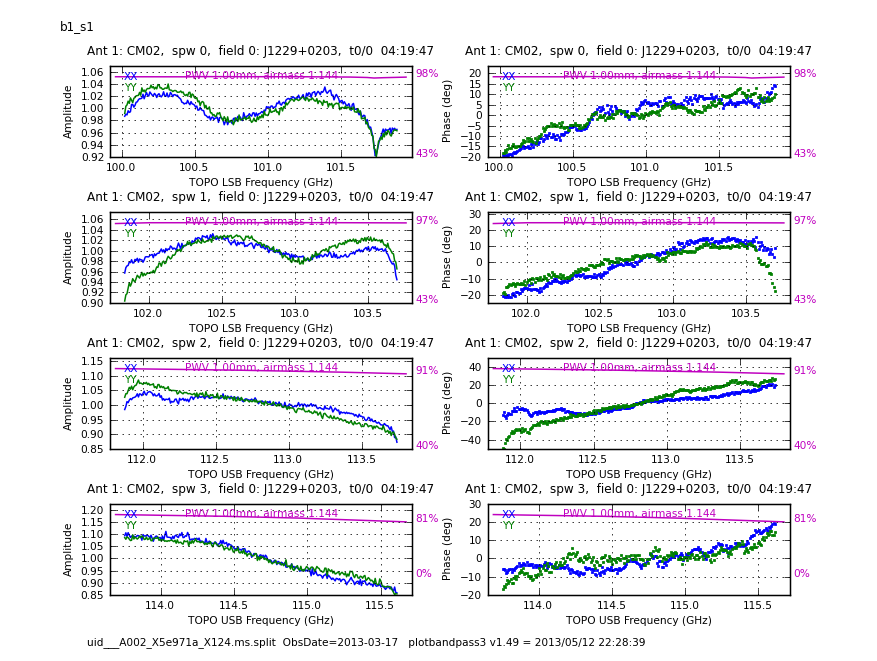
</figure>
<figure id="bandpass_freq_amp.png">
</figure>
<figure id="bandpass_freq_phase.png">
</figure>
#In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
os.system('rm -rf cal_plots/*bandpass*.png')
for name in basename:
aU.plotbandpass(caltable=name+'.bandpass.bcal',
field='',xaxis='freq',yaxis='both',
figfile='cal_plots/'+name+'.bandpass.png',
showatm=True,interactive=False,subplot=42)
Absolute Flux Calibration
The bandpass calibration will account for the phase and amplitude response of our antennas as a function of frequency. We now solve for the absolute flux scale of the data by referencing to 3c273 and in the next section we will calibrate the phase and amplitude behavior of the antennas as a function of time. However, Titan is not available for these dataset, instead we will use flux of 3c273. Next, we will use the setjy task to read the predicted complex visibilities for J1229+0203 into the MODEL column of each data set.
We will put in best flux density for 3c273 instead of Titan.
- X124, X3f3 and X5b3: we will use the value observed on March 17, 2013.
- X2e7 and X44: we will use the value on April 14, 2013.
- X4eb: the latest value will be used.
# in CASA
# Put in best flux density for 3c273 on Mar 17, 2013
setjy(vis = 'uid___A002_X5e971a_X124.ms',
field = 'J1229+0203',
fluxdensity = [8.0,0,0,0],
spw = '16,18',
usescratch = F)
setjy(vis = 'uid___A002_X5e971a_X124.ms',
field = 'J1229+0203',
fluxdensity = [7.7,0,0,0],
spw = '20,22',
usescratch = F)
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
setjy(vis = name+'.ms',
field = 'J1229+0203',
fluxdensity = [7.7,0,0,0],
spw = '8,10',
usescratch = F)
# Put in best flux density for 3c273 on Apr 14, 2013
for name in ['uid___A002_X5e971a_X2e7','uid___A002_X60b415_X44']:
setjy(vis = name+'.ms',
field = 'J1229+0203',
fluxdensity = [8.0,0,0,0],
spw = '16,18',
usescratch = F)
setjy(vis = name+'.ms',
field = 'J1229+0203',
fluxdensity = [7.7,0,0,0],
spw = '20,22',
usescratch = F)
# The latest flux density of 3c273
setjy(vis = 'uid___A002_X62f759_X4eb.ms',
field = 'J1229+0203',
spw = '16,18',
fluxdensity = [7.8,0,0,0],
usescratch = F)
setjy(vis = 'uid___A002_X62f759_X4eb.ms',
field = 'J1229+0203',
spw = '20,22',
fluxdensity = [7.4,0,0,0],
usescratch = F)
setjy will output the flux of 3c273 to the CASA logger and it is worth recording this information.
Gain (Phase and Amplitude) Calibration
With the flux now properly scaled, we will calibrate the phase and amplitude behavior of the antennas as a function of time.
We begin by running a short-solution interval gaincal to solve for phase variation on short timescales during observations of our two calibrators, with solint set to "int". By applying this on-the-fly, we can remove any decorrelation in the data due to phase scatter when we solve for the amplitude calibration. However, there is no benefit to using this short-timescale solution to calibrate the source because we only have information on the gain during calibrator visits (though see the Imaging portion of this guide). Instead we will solve for the gains to apply to the source using a longer solint in just a moment.
# in CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print "Carrying out short timescale phase solution for "+name
os.system('rm -rf '+name+'.intphase.gcal')
gaincal(vis=name+'.ms',
gaintable=name+'.bandpass.bcal',
caltable=name+'.intphase.gcal',
calmode='p',
field='J1229+0203',
spw='',
refant='CM04', solint='int',minsnr=3.0)
Now we derive the longer timescale phase calibration table using solint set to "inf", but not allowing scan combination. This calibration has higher signal to noise due to combining more data, and for the purposes of correcting the source, it is just as precise as the short timescale solution.
# in CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print "Carrying out longer timescale phase solution for "+name
os.system('rm -rf '+name+'.scanphase.gcal')
gaincal(vis=name+'.ms',
gaintable=name+'.bandpass.bcal',
caltable=name+'.scanphase.gcal',
calmode='p',
field='J1229+0203',
spw='',
refant='CM04', solint='inf',minsnr=2.0)
Now we apply the short-timescale phase solution and carry out a scan length (solint set to "inf") calibration of the data using calmode of 'ap'.
# in CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print "Solving for longer (scan) interval amplitude solution for "+name
os.system('rm -rf '+name+'.amp.cal')
gaincal(vis = name+'.ms',
gaintable =[name+'.bandpass.bcal',name+'.intphase.gcal'],
caltable = name+'.amp.cal',
calmode='ap',
field = ['J1229+0203', 'J1215+1654'],
spw='',
refant = 'CM04',solint = 'inf', minsnr=2.0)
This "amp.cal" solution gives us the amplitude variations as a function of time, but they are not yet pinned to a realistic scale except in the case of Titan, where we have solved using the model input by setjy. Here, we will set the flux of our secondary calibrator J1215+1654 with reference to 3c273 using fluxscale.
# in CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print "Scaling amplitude calibration to match 3c273 for "+name
os.system('rm -rf '+name+'.flux.cal')
fluxscale(vis = name+'.ms',
caltable = name+'.amp.cal',
fluxtable = name+'.flux.cal',
reference = 'J1229+0203',
transfer = 'J1215+1654')
This new correctly-scaled flux table ".flux.cal" replaces the previous ".amp.cal" table as the correct amplitude calibration table to apply to the data, i.e., the ".flux.cal" contains both the time variability of the amplitude solved for in ".amp.cal" and the correct flux scaling set with fluxscale.
Now we plot the final phase and amplitude calibration tables for each data set. A well-behaved calibration table will show smooth variations as a function of time. Sudden jumps or wild variations among the antenna amplitude gains should prompt further investigation and possibly additional flagging. Here, we will present the examples of X124 dataset.
# in CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename[0]:
print "Plotting solutions for "+name
plotcal(caltable = name+'.scanphase.gcal',
xaxis = 'time', yaxis = 'phase',
iteration = 'antenna', plotrange=[0,0,-180,180],
showgui=False, subplot=421, figfile=name+'.scanphase.page1.png',
antenna='', fontsize=6.0)
# dummy_string = raw_input("Hit <Enter> to see next plot.")
plotcal(caltable = name+'.scanphase.gcal',
xaxis = 'time', yaxis = 'phase',
iteration = 'antenna', plotrange=[0,0,-180,180],
showgui=False, subplot=421, figfile=name+'.scanphase.page2.png',
antenna='', fontsize=6.0)
# dummy_string = raw_input("Hit <Enter> to see next plot.")
plotcal(caltable = name+'.flux.cal',
xaxis = 'time',yaxis = 'amp',
plotrange = [0,0,0,0],
antenna='0~7', iteration='antenna',
showgui=False, subplot=421, figfile=name+'.flux.page1.png',
fontsize=6.0)
# dummy_string = raw_input("Hit <Enter> to see next plot.")
plotcal(caltable = name+'.flux.cal',
xaxis = 'time',yaxis = 'amp',
plotrange = [0,0,0,0],
antenna='', iteration='antenna', fontsize=6.0,
showgui=False, subplot=421, figfile=name+'.flux.page2.png')
# dummy_string = raw_input("Hit <Enter> to see next plot.")
Apply the Calibrations and Inspect
Now we will use applycal to apply the bandpass, phase, and amplitude calibration tables that we generated in the previous sections to the data. We apply the solutions to the target source.
#In CASA
from recipes.almahelpers import tsysspwmap
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1229+0203',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
interp = 'linear, linear',
gainfield=['J1229+0203',''],
calwt=T,
flagbackup=F)
# Datasets where the Tsys is done on M100
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X62f759_X4eb']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1215+1654,M100',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
interp = 'linear, linear',
gainfield=['M100'],
calwt=T,
flagbackup=F)
# Dataset where Tsys is done on the phase calibrator rather than M100
for name in ['uid___A002_X60b415_X44']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1215+1654,M100',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
interp = 'linear, linear',
gainfield=['J1215+1654'],
calwt=T,
flagbackup=F)
# Datasets that have observations of Titan
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'Titan',
spw='16,18,20,22',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='Titan',
interp='linear,linear',
calwt=T,
flagbackup=F)
# Datasets with 2 spws
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
tsysmap = tsysspwmap(vis = name+'.ms', tsystable = name+'.ms.tsys')
applycal(vis=name+'.ms', field = 'J1229+0203',
spw='8,10',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='J1229+0203',
interp='linear,linear',
calwt=T,
flagbackup=F)
applycal(vis=name+'.ms', field = 'Titan',
spw='8,10',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='Titan',
interp='linear,linear',
calwt=T,
flagbackup=F)
applycal(vis=name+'.ms', field = 'J1215+1654,M100',
spw='8,10',
gaintable = [name+'.ms.tsys',name+'.ms.antpos'],
spwmap = [tsysmap,[]],
gainfield='M100',
interp='linear,linear',
calwt=T,
flagbackup=F)
Here:
- vis and field specify the measurement set and field to calibrate.
- gaintable specifies the tables to apply to the data. There may be several tables applied all at once, here we apply the bandpass, phase, and flux calibrations.
- interp specifies the type of interpolation used to match calibration tables to the data. If there are several gaintables then interp may be either a single value (e.g., "linear" or "nearest") or a list of values with size equal to the list of gaintables. In the case where interp is a list, applycal will match each element to the corresponding gaintable (e.g., first gaintable with first interp parameter).
- gainfield specifies field from which to draw the calibration. As with interp, this can be a list of size matched to gaintable, so that the first element of gainfield will be the field used for the first table.
- calwt indicates that the weights, which were derived from the Tsys measurements, should also be calibrated. Note that calwt is True by default.
Once calibrations are applied, it is important to go back and inspect the calibrated data. New problematic antennas or baselines may be visible after calibration. Repeat the steps above, focusing on the CORRECTED data column. Bear in mind that for any point source calibrators we now expect to find phase scattering around zero and to find flat amplitudes as a function of u-v distance. Look for outliers and other signatures of problematic data. As a general rule, you will want to incorporate these data into your overall flagging script then rerun the whole calibration process, so that reduction is iterative. If the data only represent a minor problem, however, it may not be terribly harmful to flag them after the fact so that they do not interfere with imaging but trust that the calibrations are mostly unaffected.
<figure id="plotms_amp_time_phasecal.png">
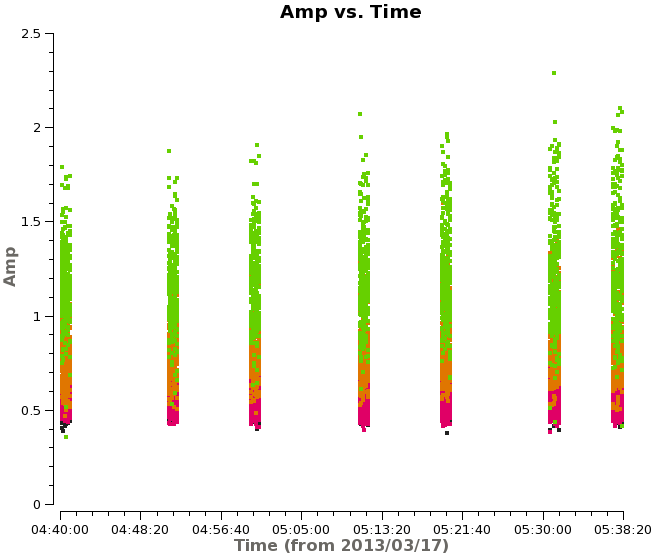
</figure>
Here, we will present the examples of X124 dataset.
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename[0] :
print "Examining calibrated data for "+name
plotms(vis = name+'.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', field='J1215+1654',
averagedata=True, avgchannel='4080', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = name+'.ms', xaxis='time', yaxis='amp',
ydatacolumn='corrected', field='J1215+1654',
averagedata=True, avgchannel='4080', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = name+'.ms', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', field='J1215+1654',avgtime='1e8',
avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = name+'.ms', xaxis='time', yaxis='phase',
ydatacolumn='corrected', field='J1215+1654',
avgchannel='4080', avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
In <xr id="plotms_amp_time_phasecal.png"/>, we plot phase vs. channel and amp vs. time for J1215+1654 for the X124 dataset.
Finally we can use plotms to examine the corrected amplitude and phase of M100 as a function of time and uv-distance (plots not shown):
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename[0]:
print "Examining calibrated data for "+name
plotms(vis = name+'.split.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', field='M100',
averagedata=True, avgchannel='4080', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = name+'.ms', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', field='M100',
avgtime='1e8', avgscan=T, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
Split and Concatenate Data for M100 Mosaic
The individual data sets are now calibrated. We can safely split out the calibrated data for our science target and drop the calibrators.
#In CASA
os.system('rm -rf *.ms.split')
for name in ['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']:
split(vis = name+'.ms',
outputvis = name+'.ms.split',
datacolumn = 'corrected',
spw = '16,18,20,22',
keepflags = F)
for name in ['uid___A002_X5e9ff1_X3f3','uid___A002_X5e9ff1_X5b3']:
split(vis = name+'.ms',
outputvis = name+'.ms.split',
datacolumn = 'corrected',
spw = '8,10',
keepflags = F)
Before we do this, we will take a brief aside to remove Pointing information from the calibrated data. Normally, the pointing information would apply important corrections to be used while imaging the data. However, some aspects of this are still under development and the pointing information in these data interacts poorly with CASA. We will remove the pointing information using the CASA toolkit Table tool (tb). This kind of operation is dangerous and should not be a regular feature of your CASA reduction.
# In CASA
basename=['uid___A002_X5e971a_X124','uid___A002_X5e971a_X2e7','uid___A002_X5e9ff1_X3f3',
'uid___A002_X5e9ff1_X5b3', 'uid___A002_X60b415_X44','uid___A002_X62f759_X4eb']
for name in basename:
print "Manually removing pointing table from "+name+".cal.ms ... Please be careful!"
tb.open(name+'.cal.ms/POINTING',nomodify=False)
a = tb.rownumbers()
tb.removerows(a)
tb.close()
# In CASA
os.system('rm -rf M100_Band3_7m_CalibratedData.ms')
concat(vis=['X2e7-cal.ms','X3f3-cal.ms','X4eb-cal.ms','X5b3-cal.ms','X44-cal.ms','X124-cal.ms'],concatvis='M100_Band3_7m_CalibratedData.ms')
Imaging Mosaics
If you are unfamiliar with the basic concepts of deconvolution and clean, pause here and review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/wilner_synthesis10.pdf
Mosaics like other kinds of images are created in the CASA task clean. To invoke mosaic mode, you simply set the parameter imagermode='mosaic'. The default subparameter ftmachine='mosaic' is then automatically set. This is a joint deconvolution algorithm that works in the uv-plane. A convolution of the primary beam patterns for each pointing in the mosaic is created: the primary beam response function. The corresponding image of the mosaic response function will be called <imagename>.flux.pbcoverage and <imagename>.flux (where the latter differs from the former only if the sensitivity of each field in the mosaic varies).
If you want to learn more about mosaicing, pause here and review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/jott-mosaicking-school-04.pdf
Continuum subtraction and Split out CO(1-0) spw
In these data, the continuum emission is too weak to contaminate the line emission (i.e. the peak continuum emission is less than the rms noise in the spectral line channels). Nevertheless, for illustrative purposes we demonstrate how to subtract the continuum emission in the uv-domain using the task uvcontsub2.
# In CASA
uvcontsub2(vis='M100_Band3_7m_CalibratedData.ms',fitspw='0;1:128~1850;2150~3951',fitorder = 1, combine = 'spw', spw = '0,1')
Here, fitspw gives the line-free channels for each mosaic and fitorder=1. The output MS will have .contsub appended to the name.
# In CASA
split(vis = 'M100_Band3_7m_CalibratedData.ms', outputvis= 'M100_7m_CO.ms', field='M100',spw='1,3', datacolumn = 'data', keepflags=F)
CO(1-0) Imaging
<figure id="M100_CO1_0_vel.png">
</figure>
Now we are ready to make cubes of the line emission. The imaging parameters are similar to the continuum except for those dealing with the spectral setup: mode, start, width, nchan, restfreq, and outframe parameters. When making spectral images you have three choices for the mode parameter: channel, velocity, and frequency. Data are taken using constant frequency channels. For spectral line analysis it's often more useful to have constant velocity channels, and this is also the best way to make images of multiple lines with the exact same channelization for later comparison. For mode='velocity', the desired start and width also need to be given in velocity units for the desired output frame.
It is important to note that ALMA does not do on-line Doppler Tracking and the native frame of the data is TOPO. If you do not specify outframe the output cube will also be in TOPO, which is not very useful for scientific analysis. The Doppler Shift is taken out during the regridding to the desired outframe in clean or alternatively it can be done separately by the cvel task which would need to be run before clean.
At first, we will make initial dirty image.
# In CASA
os.system('rm -rf M100.CO1_0_vel.png')
plotms(vis='M100_Band3_7m_CalibratedData.ms.contsub',xaxis='velocity',yaxis='amp',
avgtime='1e8',avgscan=T,transform=T,freqframe='LSRK',
restfreq='115.271201800GHz',plotfile='M100.CO1_0_vel.png')
# In CASA
# Define clean parameters
vis='M100_7m_CO.ms'
prename='M100_7m_cube'
myimage=prename+'.image'
myflux=prename+'.flux'
mymask=prename+'.mask'
myresidual=prename+'.residual'
imsize=250
cell='2.0arcsec'
minpb=0.2
restfreq='115.271201800GHz'
outframe='LSRK'
spw='0~1'
width='5km/s'
start='1400km/s'
nchan=70
robust=0.5
phasecenter='J2000 12h22m54.9 +15d49m10'
Setup stopping criteria
# In CASA
stop=3. # multiplier for rms
Minimum size multiplier for beam area for removing very small mask regions.
# In CASA
pixelmin=0.5 # reasonable default is 1/2 the beam area
# In CASA
os.system('rm -rf '+prename+'.* ' +prename+'_*')
clean(vis=vis,imagename=prename,
imagermode='mosaic',ftmachine='mosaic',minpb=minpb,
imsize=imsize,cell=cell,spw=spw,
weighting='briggs',robust=robust,phasecenter=phasecenter,
mode='velocity',width=width,start=start,nchan=nchan,
restfreq=restfreq,outframe=outframe,veltype='radio',
mask='',
niter=0,interactive=F)
Inspect the resulting data cube:
# In CASA
viewer('M100.7m.CO.image')
Find properties of the dirty image
# In CASA
myimage=prename+'.image'
bigstat=imstat(imagename=myimage)
peak= bigstat['max'][0]
print 'peak in cube = '+str(peak)
thresh = peak /2. # sets loop threshold
Determine or Set rms
In order to find properties of the dirty image, we will check the rms
# In CASA
if True:
# If True: find the rms in two line-free channels
chanstat=imstat(imagename=myimage,chans='4')
rms1= chanstat['rms'][0]
chanstat=imstat(imagename=myimage,chans='66')
rms2= chanstat['rms'][0]
rms=0.5*(rms1+rms2)
else:
# Set rms by hand
rms=0.013
Make Final Images
# In CASA
print 'rms in a channel = '+str(rms)
Deterimine the beam area in pixels for later removal of very small mask regions
# In CASA
major=imhead(imagename=myimage,mode='get',hdkey='beammajor')['value']
minor=imhead(imagename=myimage,mode='get',hdkey='beamminor')['value']
pixelsize=float(cell.split('arcsec')[0])
beamarea=(major*minor*pi/(4*log(2)))/(pixelsize**2)
print 'beamarea in pixels =', beamarea
n=-1
while (thresh >= stop*rms):
n=n+1
print 'clean threshold this loop is', thresh
threshmask = prename+'_threshmask' +str(n)
maskim = prename+'_fullmask' +str(n)
immath(imagename = [myresidual],
outfile = threshmask,
expr = 'iif(IM0 > '+str(thresh) +',1.0,0.0)',
mask=myflux+'>'+str(minpb))
if (n==0):
os.system('cp -r '+threshmask+' '+maskim+'.pb')
print 'This is the first loop'
else:
makemask(mode='copy',inpimage=myimage,
inpmask=[threshmask,mymask],
output=maskim)
imsubimage(imagename=maskim, mask=myflux+'>'+str(minpb),
outfile=maskim+'.pb')
print 'Combined mask ' +maskim+' generated.'
os.system('cp -r '+maskim+'.pb ' +maskim+'.pb.min')
maskfile=maskim+'.pb.min'
ia.open(maskfile)
mask=ia.getchunk()
labeled,j=scipy.ndimage.label(mask)
myhistogram = scipy.ndimage.measurements.histogram(labeled,0,j+1,j+1)
object_slices = scipy.ndimage.find_objects(labeled)
threshold=beamarea*pixelmin
for i in range(j):
if myhistogram[i+1]<threshold:
mask[object_slices[i]] = 0
ia.putchunk(mask)
ia.done()
print 'Small masks removed and ' +maskim +'.pb.min generated.'
os.system('rm -rf '+mymask+'')
clean(vis=vis,imagename=prename,
imagermode='mosaic',ftmachine='mosaic',minpb=minpb,
imsize=imsize,cell=cell,spw=spw,
weighting='briggs',robust=robust,phasecenter=phasecenter,
mode='velocity',width=width,start=start,nchan=nchan,
restfreq=restfreq,outframe=outframe,veltype='radio',
mask = maskim+'.pb.min',
interactive = F,
niter = 10000,
threshold = str(thresh) +'Jy/beam')
if thresh==stop*rms: break
thresh = thresh/2.
# Run a final time with stop*rms if more than a little above stop*rms
if (thresh < stop*rms and thresh*2.>1.05*stop*rms):
thresh=stop*rms
os.system('cp -r '+myimage+' '+myimage+str(n))
Image Analysis : Moment Maps
<figure id="M100.7m.CO1_0_Line.Clean.image.mom0.png">
</figure> <figure id="M100.7m.CO1_0_Line.Clean.image.mom1.png">
</figure>
Next we will make moment maps for the CO(1-0) emission: Moment 0 is the integrated intensity; Moment 1 is the intensity weighted velocity field; and Moment 2 is the intensity weighted velocity dispersion.
Above we determined the rms noise levels for M100 mosaics in a line-free and a line-bright channel. We want to limit the channel range of the moment calculations to those channels with significant emission. One good way to do this is to open the cube in the viewer overlaid with 3-sigma contours, with sigma corresponding to the line-free rms.
# In CASA
imview(raster={'file': 'M100.7m.CO1_0.image',
'range': [-0.04,0.4]},
contour={'file': 'M100.7m.CO1_0.image',
'levels': [0.004],'unit': 5})
We find a channel range for significant emission of 10~61.
For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity.
# In CASA
myimage='M100_7m_cube.image'
chanstat=imstat(imagename=myimage,chans='4')
rms1= chanstat['rms'][0]
chanstat=imstat(imagename=myimage,chans='66')
rms2= chanstat['rms'][0]
rms=0.5*(rms1+rms2)
print 'rms in a channel = '+str(rms)
os.system('rm -rf M100_7m_cube.image.mom*')
immoments(imagename = 'M100_7m_cube.image',
moments = [0],
axis = 'spectral',
chans = '10~61',
box='65,50,195,182',
includepix = [rms*2,100],
outfile = 'M100_7m_cube.image.mom0')
For higher order moments it is very important to set a conservative flux threshold. Typically something like 6sigma, using sigma from a bright line channel works well. We do this with the mask parameter in the commands below. When making multiple moments, immoments appends the appropriate file name suffix to the value of outfile.
# In CASA
immoments(imagename = 'M100_7m_cube.image',
moments = [1],
axis = 'spectral',
chans = '10~61',
box='65,50,195,182',
includepix = [rms*5.5,100],
outfile = 'M100_7m_cube.image.mom1')
Next we can see these moment maps from these images using imview.
# In CASA
os.system('rm -rf M100_7m_cube.mom*.png')
Im0 = 'M100_7m_cube.image.mom0'
Im1 = 'M100_7m_cube.image.mom1'
imview(raster = {'file': Im0,
'colorwedge': T,
'colormap': 'Rainbow 2',
'scaling': -1.6,
'range': [0.1,140],
'beamellipse':T},
out = 'M100_7m_cube.mom0.png')
imview(raster = {'file': Im1,
'colorwedge': T,
'colormap': 'Rainbow 2'},
contour = {'file': Im0,
'levels': [5,15,45,135],
'base': 0,
'unit': 1.0},
out = 'M100_7m_cube.mom1.png')
Export data as fits
If you want to analyze the data using another software package it is easy to convert from CASA format to FITS.
# In CASA
os.system('rm -rf M100_7m_*.fits')
exportfits(imagename='M100_7m_cube.image', fitsimage='M100_7m_cube.image.fits')
exportfits(imagename='M100_7m_cube.image.mom0', fitsimage='M100_7m_mom0.fits')
exportfits(imagename='M100_12m_cube.flux', fitsimage='M100_12m_cube.flux.fits')
Although "FITS format" is supposed to be a standard, in fact most packages expect slightly different things from a FITS image. If you are having difficulty, try setting velocity=T and/or dropstokes=T.
Continue on to Combining Images with SD and 12m dataset
Now you can continue on to the M100_Band3_Combine_4.1.
Last checked on CASA Version 4.1.0.