Simulation Guide for New Users (CASA 4.0): Difference between revisions
| (16 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Simulations Intro}} | {{Simulations Intro 4.0}} | ||
* '''This guide is applicable to CASA version 4.0. For older versions of CASA see [[Simulation Guide for New Users (CASA 3.4)]].''' | * '''This guide is applicable to CASA version 4.0. For older versions of CASA see [[Simulation Guide for New Users (CASA 3.4)]].''' | ||
* '''To create a script of the Python code on this page see [[Extracting scripts from these tutorials]].''' | * '''To create a script of the Python code on this page see [[Extracting scripts from these tutorials]].''' | ||
| Line 65: | Line 65: | ||
====Angular Scale==== | ====Angular Scale==== | ||
If you open the FITS image of 30 Doradus in your favorite viewer (e.g. '''viewer''' in CASA), you will see that it covers quite a large footprint on the sky, about 10' on a side. Observing such a large area would require | If you open the FITS image of 30 Doradus in your favorite viewer (e.g. '''viewer''' in CASA), you will see that it covers quite a large footprint on the sky, about 10' on a side. Observing such a large area with ALMA would require a mosaic with many pointings. To make the simulation more manageable, we are going to tell simobserve to rescale the pixels to shrink the image by roughly a factor of 15 (from 2.3" to 0.15" pixels) using the '''incell''' parameter. The model will then be approximately 40" on a side. This rescaled model will fit within a small ALMA mosaic of 6 pointings. Although we do this primarily for convenience in this example, a scientific motivation for this type of rescaling would be to approximate what a super-giant HII region like 30 Doradus would look like if moved from the Large Magellanic Cloud to the distance of M33 or M31. For the sake of demonstration, we will also change the coordinates of the center of the map (using the '''indirection''' parameter). Units are case sensitive, so please take care to enter them in verbatim. CASA will throw an error otherwise. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 75: | Line 75: | ||
====Observed Wavelength==== | ====Observed Wavelength==== | ||
The model image of 30 Doradus shows 8 micron continuum emission. ALMA does not observe at wavelengths this short, so we will tell simobserve | The model image of 30 Doradus actually shows 8 micron continuum emission. ALMA does not observe at wavelengths this short, so we will tell simobserve to interpret this data as if it were a 230 GHz (1.3 mm) continuum map observed with a 2 GHz bandwidth. Although for this particular example the bandwidth value is not critical, it would be very important if we were modifying a spectral cube instead of a continuum image. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 85: | Line 85: | ||
====Brightness Scale==== | ====Brightness Scale==== | ||
The 8 | The 8 micron brightness from 30 Doradus is probably not a great approximation for the millimeter brightness. For a true science case, one would want to estimate the expected brightness at 230 GHz more carefully. For the sake of demonstration, we will simply adjust the flux scale in the image so that the brightest pixel in the map has a flux density of 0.06 mJy. This number is chosen such that the fainter, extended emission in the image is a factor of a few brighter than the expected noise in a 2-hour observation. The [https://almascience.nrao.edu/call-for-proposals/sensitivity-calculator ALMA sensitivity calculator] can be used to determine the expected noise for an observation. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 92: | Line 92: | ||
</source> | </source> | ||
Remember that you can check your task inputs at any time by typing <tt>inp</tt> at the | Remember that you can check your task inputs at any time by typing <tt>inp</tt> at the CASA prompt. | ||
===Defining the Mock Observations=== | ===Defining the Mock Observations=== | ||
| Line 100: | Line 100: | ||
====Pointings and Scan Time==== | ====Pointings and Scan Time==== | ||
We will first change the | We will first change the subparameters under '''setpointings'''. | ||
The default value for '''integration''', 10 seconds, might be appropriate to simulate real observations. However, simobserve will run much faster if | The default value for '''integration''', 10 seconds, might be appropriate to simulate real observations. However, simobserve will run much faster if '''integration''' is set to a larger value, thereby reducing the number of data points. In this demonstration we will set it to 600 seconds. When simobserve is used for scientific purposes it may be best to make an initial test run with '''integration''' set to a large value, and then decrease it to a more realistic time for the final run. You will get a more accurate simulation with 10 second integrations than with 600 second integrations, especially when simulating Early Science observations with a limited number of baselines. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 109: | Line 109: | ||
</source> | </source> | ||
Note that the integration time is different than the total (on-source) time of the observations. The integration time, set here, is the averaging time for each data point. The total time spent on-source | Note that the integration time is different than the total (on-source) time of the observations. The integration time, set here, is the averaging time for each data point. The total time spent on-source can be set with the '''totaltime' parameter, as described below. Each pair of antennas will generate a number of data points equal to the total observing time divided by the integration time. | ||
We will keep '''direction''' at the default (blank) value to center the observations on the model coordinates, as given in '''indirection''' above. | We will keep '''direction''' at the default (blank) value to center the observations on the model coordinates, as given in '''indirection''' above. | ||
We will keep '''mapsize''' at the default value so that the mosaic will automatically cover the entire image. In our case, this will require a mosaic of 6 pointings | We will keep '''mapsize''' at the default value so that the mosaic will automatically cover the entire image. In our case, this will require a mosaic of 6 pointings. One could also set an exact output image size via '''mapsize = ['10arcmin','10arcmin']''' for example. | ||
The default mosaic pattern, '''maptype = 'ALMA' ''', tells simobserve to use the same hexagonal algorithm as the ALMA OT. We will leave this unchanged. | The default mosaic pattern, '''maptype = 'ALMA' ''', tells simobserve to use the same hexagonal algorithm as the ALMA OT. We will leave this unchanged. | ||
| Line 121: | Line 121: | ||
====Antenna Positions and Total Observation Time==== | ====Antenna Positions and Total Observation Time==== | ||
We will now consider the subparameters available when ''' | We will now consider the subparameters available when '''obsmode = 'int' '''. We will keep the default values for every parameter (including '''totaltime = 7200s''') except '''antennalist''', which tells simobserve the locations and sizes of each antenna in the array. We will simulate an Early Science Cycle 1 observation using a pre-defined ALMA configuration file stored in CASA. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 128: | Line 128: | ||
</source> | </source> | ||
This command sets the '''antennalist''' parameter to one of the [[Antenna_Configurations_Models_in_CASA| Cycle 1 configurations]] | This command sets the '''antennalist''' parameter to one of the [[Antenna_Configurations_Models_in_CASA| Cycle 1 configurations]]. | ||
====Thermal Noise==== | ====Thermal Noise==== | ||
| Line 147: | Line 145: | ||
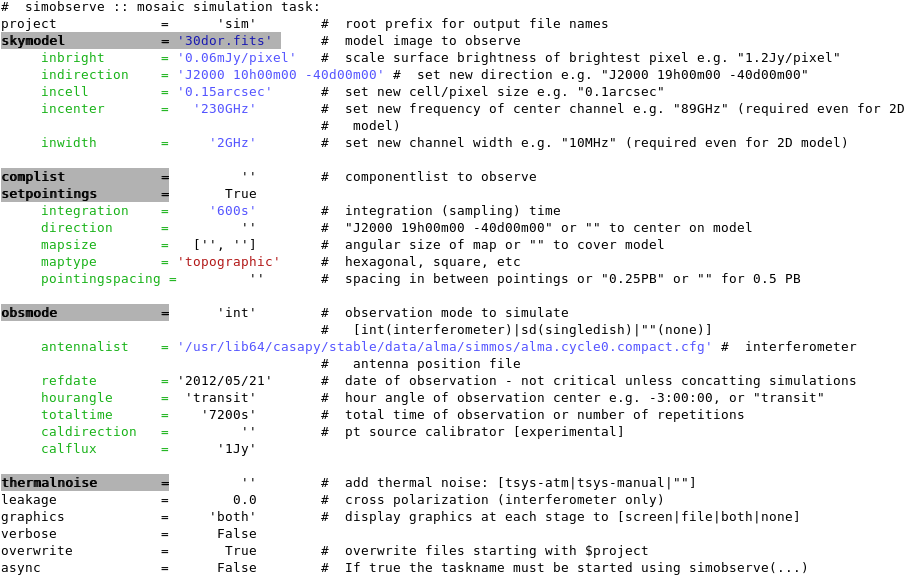
All simobserve output will be written to a directory whose name was given by the '''project''' parameter, in our case, '''project = 'sim' '''. Inside this directory, you will find: | All simobserve output will be written to a directory whose name was given by the '''project''' parameter, in our case, '''project = 'sim' '''. Inside this directory, you will find: | ||
[[File:Sim.alma cycle1 1.observe.png|thumb|right|The 2x2 summary plot output by simobserve showing source elevation vs. time, antenna position, uv coverage, and the point spread function.]] | [[File:Sim.alma cycle1 1.observe.png|thumb|right|The 2x2 summary plot output by simobserve showing source elevation vs. time, antenna position, uv coverage, and the point spread function.]] | ||
# The simulated measurement set (<tt>sim.alma_cycle1_1.ms</tt>), | # The simulated data, stored as a "measurement set" (<tt>sim.alma_cycle1_1.ms</tt>), | ||
# CASA image of the point spread function (<tt>sim.alma_cycle1_1.quick.psf</tt>), | # CASA image of the point spread function (<tt>sim.alma_cycle1_1.quick.psf</tt>), | ||
# CASA image of the input sky model rescaled according to the '''skymodel''' sub-parameters (<tt>sim.alma_cycle1_1.skymodel</tt>), | # CASA image of the input sky model rescaled according to the '''skymodel''' sub-parameters (<tt>sim.alma_cycle1_1.skymodel</tt>), | ||
| Line 162: | Line 160: | ||
==Using simanalyze== | ==Using simanalyze== | ||
Now that simobserve has created a simulated set | Now that simobserve has created a simulated data set with visibility measurements, we are ready to image the visibilities and analyze the result. The task '''simanalyze''' performs the imaging and generates useful plots and figures for analysis. We begin by resetting the simanalyze inputs to their default values. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 175: | Line 173: | ||
We want to make a deconvolved output image, but we don't want to spend too much time optimizing the cleaning. So, all we need to do is make sure the '''image''' parameter is set to True (which it is by default) and leave all of its sub-parameters at their default values. Other [http://casaguides.nrao.edu/index.php?title=Main_Page data reduction guides] describe the process of imaging in greater detail. | We want to make a deconvolved output image, but we don't want to spend too much time optimizing the cleaning. So, all we need to do is make sure the '''image''' parameter is set to True (which it is by default) and leave all of its sub-parameters at their default values. Other [http://casaguides.nrao.edu/index.php?title=Main_Page data reduction guides] describe the process of imaging in greater detail. | ||
Note that simanalyze is slightly picky about the image size. It does not like the default image size (set equal to the FITS image size) of 251. Set it to 252 | Note that simanalyze is slightly picky about the image size. It does not like the default image size (set equal to the FITS image size) of 251. Set it to 252 to make simanalyze happy. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 182: | Line 180: | ||
</source> | </source> | ||
For simulations intended for a proposal or scientific analysis, one would almost certainly want to choose a more appropriate cleaning threshold and define the region to be cleaned. Instructions for how to define the region to be cleaned with the | For simulations intended for a proposal or scientific analysis, one would almost certainly want to choose a more appropriate cleaning threshold and define the region to be cleaned. Instructions for how to define the region to be cleaned with the '''mask''' parameter can be found by typing | ||
> help clean | > help clean | ||
or by looking at the [http://casaguides.nrao.edu/index.php?title=Clean Clean CASA Guide page]. | or by looking at the [http://casaguides.nrao.edu/index.php?title=Clean Clean CASA Guide page]. | ||
| Line 223: | Line 221: | ||
All CASA images written to disk can be opened later using the CASA viewer. Just type | All CASA images written to disk can be opened later using the CASA viewer. Just type | ||
> viewer | > viewer | ||
at the CASA prompt to start the viewer tool, and navigate to the | at the CASA prompt to start the viewer tool, and navigate to the <tt>sim</tt> directory. To display the simulated observations, first select <tt>sim.alma_cycle1_1.image</tt>, then click the "raster image" button. The image will be shown in the viewer. Clicking on the picture of the wrench in the upper left corner of the Viewer Display Panel will open a window with data display options, such as changing the color scale, changing the coordinate scale, and modifying the axis labels. The image to the right was created by: | ||
# changing <tt>basic settings -> color map</tt> from "Rainbow 2" to "Hot Metal 1", | # changing <tt>basic settings -> color map</tt> from "Rainbow 2" to "Hot Metal 1", | ||
| Line 229: | Line 227: | ||
# and changing <tt>color wedge -> display color wedge</tt> from "No" to "Yes". | # and changing <tt>color wedge -> display color wedge</tt> from "No" to "Yes". | ||
{{Simulations Intro 4.0}} | |||
{{Simulations Intro}} | |||
[[Category: Simulations]] [[Category:ALMA]] | [[Category: Simulations]] [[Category:ALMA]] | ||
{{Checked 4.0.0}} | {{Checked 4.0.0}} | ||
Latest revision as of 19:03, 2 November 2012
↵ Simulating Observations in CASA
- This guide is applicable to CASA version 4.0. For older versions of CASA see Simulation Guide for New Users (CASA 3.4).
- To create a script of the Python code on this page see Extracting scripts from these tutorials.
Explanation of the guide
When planning an interferometric observation it is useful to simulate the output of the instrument under a variety of conditions. In CASA, interferometric observations can be simulated using task simobserve and quickly analyzed using task simanalyze. This guide will demonstrate how to simulate an ALMA observation using these tasks. (Task simobserve can also be used to simulate observations with other interferometers, but this currently requires advanced techniques not covered here.)
We begin with an image similar to something that might be observed with ALMA. We show how to rescale the image and specify the ALMA antenna configuration. We produce a simulated set of visibility measurements and then produce an image from the visibility data. In the process, we also generate useful figures to help us analyze the simulated output.
We will assume throughout only a general knowledge of interferometry and no specific knowledge of CASA.
Getting Started
The two things you need to get started are
- The image we will work with, and
- CASA version 4.0.
We will use the Spitzer IRAC 8 micron image of 30 Doradus, which you can download here. See the Simulation Inputs page for other sample images.
To install CASA, follow the instructions given on the Obtaining CASA page.
CASA Basics
CASA is the post-processing package for ALMA and EVLA and can handle both interferometric and single dish data. Because simobserve and simanalyze are tasks within CASA, we start here with a brief introduction to some CASA basics. To learn much more about CASA, go to the CASA homepage. Walk-throughs of CASA data reduction for a variety of data sets can be found on the CASA Guides website.
Once you have installed CASA, you can launch it by typing "casapy" at the prompt or by double-clicking on the icon, depending on your system and preferences.
To see a list of all available CASA tasks, at the CASA prompt type
tasklist
To look at the inputs for an available task, use "inp". For instance
inp simobserve
Any parameter with a grey background is expandable. Black text shows a default value. Red text shows an invalid value. Blue text shows a valid value different from the default.
To get help on a given task, use "help" (to exit from help, hit the "q" key). For instance
help simanalyze
To reset a task to its default values, use "default". For instance
default simobserve
To run a task using the current global values of its input parameters, just type its name at the CASA prompt. For instance
simobserve
It's a good idea to double-check the input parameters (i.e., re-run inp simobserve) immediately before running a task.
Using simobserve
The task simobserve uses a model image along with a number of input parameters that define the observing conditions and alter the input model. The output of simobserve is a "visibility measurement set" (i.e. the simulated raw data in CASA format) and various diagnostic plots and images.
Getting Your Input Image Into simobserve
First, we'll tell simobserve where to find the model (input) image. Next we will rescale the image to meet our purposes. Just to be safe, we'll begin by restoring the default values of simobserve and then set the 30 Doradus image as the input model using the skymodel parameter. Note that you need to use the full path to your model image if it is not in your current working directory.
#In CASA
default simobserve
skymodel = '30dor.fits'
Once skymodel is set, the command inp reveals sub-parameters that control scaling the model image.
Angular Scale
If you open the FITS image of 30 Doradus in your favorite viewer (e.g. viewer in CASA), you will see that it covers quite a large footprint on the sky, about 10' on a side. Observing such a large area with ALMA would require a mosaic with many pointings. To make the simulation more manageable, we are going to tell simobserve to rescale the pixels to shrink the image by roughly a factor of 15 (from 2.3" to 0.15" pixels) using the incell parameter. The model will then be approximately 40" on a side. This rescaled model will fit within a small ALMA mosaic of 6 pointings. Although we do this primarily for convenience in this example, a scientific motivation for this type of rescaling would be to approximate what a super-giant HII region like 30 Doradus would look like if moved from the Large Magellanic Cloud to the distance of M33 or M31. For the sake of demonstration, we will also change the coordinates of the center of the map (using the indirection parameter). Units are case sensitive, so please take care to enter them in verbatim. CASA will throw an error otherwise.
#In CASA
incell = '0.15arcsec'
indirection = 'J2000 10h00m00 -40d00m00'
Observed Wavelength
The model image of 30 Doradus actually shows 8 micron continuum emission. ALMA does not observe at wavelengths this short, so we will tell simobserve to interpret this data as if it were a 230 GHz (1.3 mm) continuum map observed with a 2 GHz bandwidth. Although for this particular example the bandwidth value is not critical, it would be very important if we were modifying a spectral cube instead of a continuum image.
#In CASA
incenter = '230GHz'
inwidth = '2GHz'
Brightness Scale
The 8 micron brightness from 30 Doradus is probably not a great approximation for the millimeter brightness. For a true science case, one would want to estimate the expected brightness at 230 GHz more carefully. For the sake of demonstration, we will simply adjust the flux scale in the image so that the brightest pixel in the map has a flux density of 0.06 mJy. This number is chosen such that the fainter, extended emission in the image is a factor of a few brighter than the expected noise in a 2-hour observation. The ALMA sensitivity calculator can be used to determine the expected noise for an observation.
#In CASA
inbright = '0.06mJy/pixel'
Remember that you can check your task inputs at any time by typing inp at the CASA prompt.
Defining the Mock Observations
Now that simobserve knows how to interpret the input image, the next step is to define the simulated observations.
Pointings and Scan Time
We will first change the subparameters under setpointings.
The default value for integration, 10 seconds, might be appropriate to simulate real observations. However, simobserve will run much faster if integration is set to a larger value, thereby reducing the number of data points. In this demonstration we will set it to 600 seconds. When simobserve is used for scientific purposes it may be best to make an initial test run with integration set to a large value, and then decrease it to a more realistic time for the final run. You will get a more accurate simulation with 10 second integrations than with 600 second integrations, especially when simulating Early Science observations with a limited number of baselines.
# In CASA
integration = '600s'
Note that the integration time is different than the total (on-source) time of the observations. The integration time, set here, is the averaging time for each data point. The total time spent on-source can be set with the totaltime' parameter, as described below. Each pair of antennas will generate a number of data points equal to the total observing time divided by the integration time.
We will keep direction at the default (blank) value to center the observations on the model coordinates, as given in indirection above.
We will keep mapsize at the default value so that the mosaic will automatically cover the entire image. In our case, this will require a mosaic of 6 pointings. One could also set an exact output image size via mapsize = ['10arcmin','10arcmin'] for example.
The default mosaic pattern, maptype = 'ALMA' , tells simobserve to use the same hexagonal algorithm as the ALMA OT. We will leave this unchanged.
Finally, we will also leave pointingspacing to its default value (blank), which automatically sets the pointings to be half a primary beam apart, corresponding to Nyquist sampling.
Antenna Positions and Total Observation Time
We will now consider the subparameters available when obsmode = 'int' . We will keep the default values for every parameter (including totaltime = 7200s) except antennalist, which tells simobserve the locations and sizes of each antenna in the array. We will simulate an Early Science Cycle 1 observation using a pre-defined ALMA configuration file stored in CASA.
#In CASA
antennalist = "alma_cycle1_1.cfg"
This command sets the antennalist parameter to one of the Cycle 1 configurations.
Thermal Noise
For this simple simulation, we will not include any thermal noise in the observations, so we can leave thermalnoise at its default (empty string) value.
simobserve Execution and Output
With all the input parameters set, we are ready to execute the task:
#In CASA
simobserve()
All simobserve output will be written to a directory whose name was given by the project parameter, in our case, project = 'sim' . Inside this directory, you will find:
- The simulated data, stored as a "measurement set" (sim.alma_cycle1_1.ms),
- CASA image of the point spread function (sim.alma_cycle1_1.quick.psf),
- CASA image of the input sky model rescaled according to the skymodel sub-parameters (sim.alma_cycle1_1.skymodel),
- flattened CASA image of the input sky model rescaled (sim.alma_cycle1_1.skymodel.flat),
- flattened PNG image of the input sky model rescaled and overlaid with the mosaic pattern specified by the setpointings sub-parameters (sim.alma_cycle1_1.skymodel.png),
- a 2x2 PNG summary plot (sim.alma_cycle1_1.observe.png) showing,
- source elevation vs. time,
- antenna position,
- uv coverage,
- and the point spread function,
- and an ASCII text listing of mosaic pointings (sim.alma_cycle1_1.ptg.txt).
When simobserve is executed, the 2x2 PNG plot will be displayed in the CASA plotter.
Using simanalyze
Now that simobserve has created a simulated data set with visibility measurements, we are ready to image the visibilities and analyze the result. The task simanalyze performs the imaging and generates useful plots and figures for analysis. We begin by resetting the simanalyze inputs to their default values.
#In CASA
default simanalyze
We next setup the imaging and analysis input parameters.
Image
We want to make a deconvolved output image, but we don't want to spend too much time optimizing the cleaning. So, all we need to do is make sure the image parameter is set to True (which it is by default) and leave all of its sub-parameters at their default values. Other data reduction guides describe the process of imaging in greater detail.
Note that simanalyze is slightly picky about the image size. It does not like the default image size (set equal to the FITS image size) of 251. Set it to 252 to make simanalyze happy.
#In CASA
imsize = 252
For simulations intended for a proposal or scientific analysis, one would almost certainly want to choose a more appropriate cleaning threshold and define the region to be cleaned. Instructions for how to define the region to be cleaned with the mask parameter can be found by typing
> help clean
or by looking at the Clean CASA Guide page.
Analyze
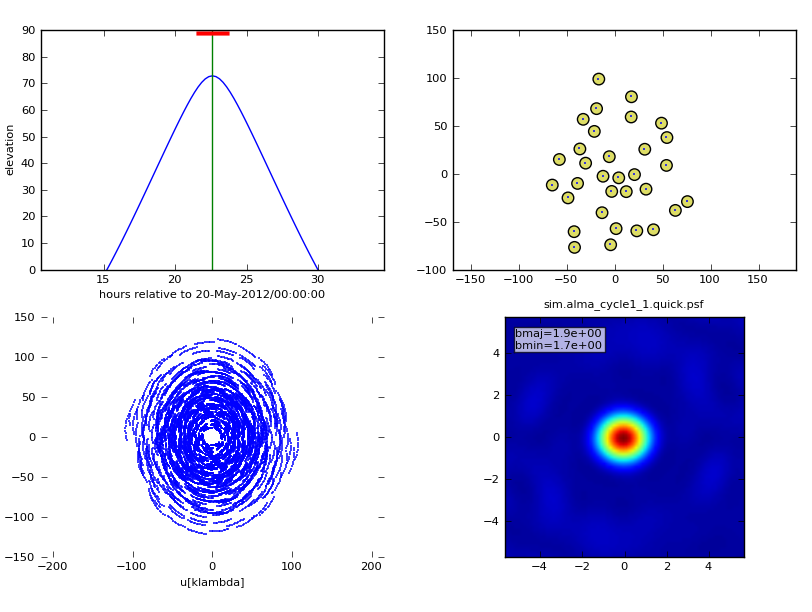
To specify how simanalyze displays the imaged data, set the parameter analyze to True and then pick your favorite output formats. If the graphics parameter is set to 'screen' or 'both', up to six output formats will be displayed in the CASA plotter. More than six outputs can be written to disk (graphics = 'file' or 'both'), but only six can be displayed in the plotter. In this example we will look at,
- the uv coverage in the 2 hour observation,
- the synthesized ("dirty") beam (point spread function),
- the original sky model (as defined in "modifymodel"),
- the convolved model (sky model convolved with the output "clean" beam),
- the clean image (the sky as observed with the interferometer after deconvolution),
- and the difference between the clean image and the convolved model.
To make these choices, use the following lines in CASA
#In CASA
analyze = True
showconvolved = True
showfidelity = False
simanalyze Execution and Output
Execute simanalyze by typing,
#In CASA
simanalyze()
The six outputs we selected will be displayed in the CASA plotter and written to disk in the sim directory.
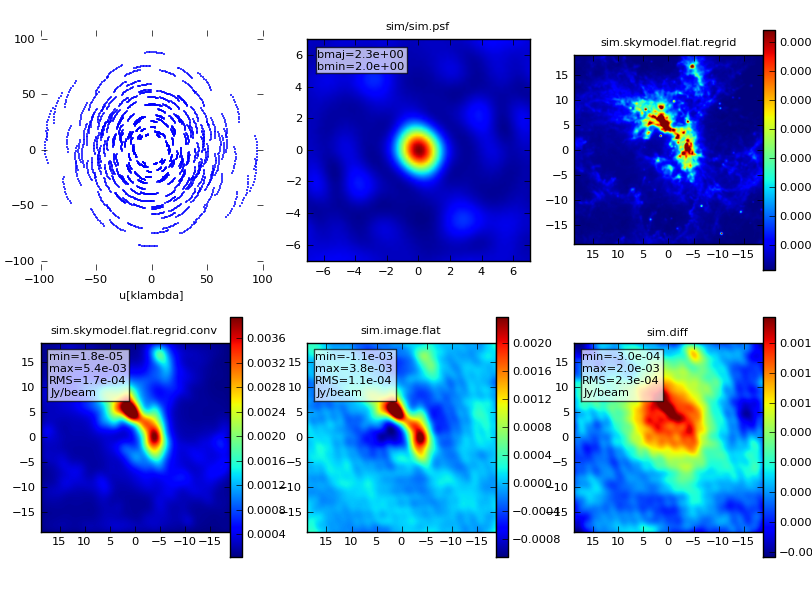
All CASA images written to disk can be opened later using the CASA viewer. Just type
> viewer
at the CASA prompt to start the viewer tool, and navigate to the sim directory. To display the simulated observations, first select sim.alma_cycle1_1.image, then click the "raster image" button. The image will be shown in the viewer. Clicking on the picture of the wrench in the upper left corner of the Viewer Display Panel will open a window with data display options, such as changing the color scale, changing the coordinate scale, and modifying the axis labels. The image to the right was created by:
- changing basic settings -> color map from "Rainbow 2" to "Hot Metal 1",
- changing beam ellipse -> beam style from "outline" to "filled",
- and changing color wedge -> display color wedge from "No" to "Yes".
↵ Simulating Observations in CASA
Last checked on CASA Version 4.0.0.