Guide to the NA Imaging Template: Difference between revisions
| (30 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
== About this Guide == | == About this Guide == | ||
This guide describes the template | This guide describes the template scripts used to manually image data for quality assurance by the North America ALMA Science Center (NAASC). This script is designed to guide the user through the decisions needed when imaging ALMA data. It should not be applied blindly; some editing of the scripts will be required in almost all cases. Not all sections of the template are relevant for all data sets. It can be adapted for use with partial pipeline processing of images. For example, you could do continuum subtraction in the pipeline, but use the scripts here as a template for manual imaging. See the [[ALMA_Imaging_Pipeline_Reprocessing | ALMA Imaging Pipeline Reprocessing]] guide for information on pipeline processed images. | ||
A current version of the script can be found on [https://github.com/aakepley/ALMAImagingScript github]. A description of how the script is used at the NAASC can be found in [http://adsabs.harvard.edu/abs/2016SPIE.9910E..0CK Kepley et al. Proceedings of the SPIE, 9910, 99100C ] | A current version of the script can be found on [https://github.com/aakepley/ALMAImagingScript github]. A description of how the script is used at the NAASC can be found in [http://adsabs.harvard.edu/abs/2016SPIE.9910E..0CK Kepley et al. Proceedings of the SPIE, 9910, 99100C ]. | ||
More information on the ALMA Quality Assurance procedure and standards can be found on the [https://almascience.org/processing/qa2-data-products ALMA Science Portal]. | |||
All example images were produced using ALMA Science Verification Data of TW Hya. More information along with the dataset can be obtained from the [https://casaguides.nrao.edu/index.php/TWHydraBand7 TW Hydra Band 7 Calibration and Imaging Guide]. | All example images were produced using ALMA Science Verification Data of TW Hya. More information along with the dataset can be obtained from the [https://casaguides.nrao.edu/index.php/TWHydraBand7 TW Hydra Band 7 Calibration and Imaging Guide]. | ||
* [[First_Look_at_Imaging | Continuum Imaging]] | |||
* [[First_Look_at_Self_Calibration | Continuum Self-Calibration]] | |||
* [[First_Look_at_Line_Imaging | Spectral Line Imaging]] | |||
* [[First_Look_at_Image_Analysis | Moments and Image Analysis]] | |||
== Prepare for Imaging == | == Prepare for Imaging == | ||
Before you begin imaging, | Before you begin imaging, determine whether the dataset was manually or pipeline calibrated. The README file should state how the dataset was calibrated. If it does not, look in the script directory of the delivered data package. If you see a file named <ASDM UID>.ms.scriptForCalibration.py, the dataset was manually calibrated. If you see a filename PPR_*.xml, the dataset was pipeline calibrated. | ||
If the data was calibrated prior to CASA 4.3, we recommend that you image the data in the same version of CASA that it was calibrated in because the weighting scheme for the data in CASA was in | If the data was calibrated prior to CASA 4.3, we recommend that you image the data in the same version of CASA that it was calibrated in because the weighting scheme for the data in CASA was in limbo. See [https://casaguides.nrao.edu/index.php/DataWeightsAndCombination Data Weights and Combination] for more information. If the data was calibrated in CASA version 4.3 or later, imaging the data in a later version of CASA should work but we recommend using the same version of Casa used for calibration when imaging since Casa is not guaranteed to be backwards compatible. The README or CASA log file will indicate which CASA version was used to calibrate the data. Different versions of CASA can be obtained from [https://casa.nrao.edu/casa_obtaining.shtml Download CASA]. Remember to check the operating system requirements before proceeding with the installation. | ||
Instructions on restoring the calibrated measurement set should be found in the README of your delivered dataset. For most cases, this can be done by executing scriptForPI.py in CASA | Instructions on restoring the calibrated measurement set should be found in the README of your delivered dataset. For most cases, this can be done by running CASA in the script directory and executing scriptForPI.py in CASA: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 22: | Line 29: | ||
== Attached Scripts == | == Attached Scripts == | ||
Two template scripts are available for this guide. These contain commands with minimal instructions and are intended to be used by those more familiar with imaging. | Two template scripts (scriptForImagingPrep_template.py and scriptForImaging_template.py) are available for this guide. These contain commands with minimal instructions and are intended to be used by those more familiar with imaging. | ||
A current version of the scripts can be found on [https://github.com/aakepley/ALMAImagingScript github]. | |||
== Obtaining Analysis Utilities == | == Obtaining Analysis Utilities == | ||
| Line 32: | Line 38: | ||
== Background Information == | == Background Information == | ||
This guide assumes that you are familiar with basic Python. If you need more information about python, see the official [https://docs.python.org/ | This guide assumes that you are familiar with basic Python. If you need more information about python, see the official [https://docs.python.org/3/tutorial/index.html Python 3.13 tutorial] | ||
If you are new to interferometry or CASA, we recommend you follow the [https://casaguides.nrao.edu/index.php?title=ALMAguides “First Look” Imaging Tutorials]. The [https:// | If you are new to interferometry or CASA, we recommend you follow the [https://casaguides.nrao.edu/index.php?title=ALMAguides “First Look” Imaging Tutorials]. The [https://web.cvent.com/event/90ae72df-7675-41b5-b056-48af11e1aa7f/summary Synthesis Imaging Summer School Lectures] provide a wealth of information on interferometry. | ||
In addition, many frequently asked questions can be found in the [https://help.almascience.org/index.php?/default/Knowledgebase/List ALMA Knowledgebase]. If you have additional questions after reviewing this guide, please contact the [https://help.almascience.org ALMA Helpdesk]. | In addition, many frequently asked questions can be found in the [https://help.almascience.org/index.php?/default/Knowledgebase/List ALMA Knowledgebase]. If you have additional questions after reviewing this guide, please contact the [https://help.almascience.org ALMA Helpdesk]. | ||
| Line 42: | Line 48: | ||
This guide has been split into 4 sections: | This guide has been split into 4 sections: | ||
#'''[[Imaging_Prep | Prepare the data for Imaging]]''' This page creates the calibrated_final.ms that will be used for imaging. Commands can be found in scriptForImagingPrep_template.py | #'''[[Imaging_Prep | Prepare the data for Imaging]]''' This page creates the calibrated_final.ms that will be used for imaging. Commands can be found in scriptForImagingPrep_template.py available on [https://github.com/aakepley/ALMAImagingScript github]. | ||
#'''[[Image_Continuum | Image the Continuum]]''' This page creates a continuum measurement set (MS) and continuum images using the MS created in [[Imaging_Prep | Prepare the data for imaging]] section. Commands can be found in | #'''[[Image_Continuum | Image the Continuum]]''' This page creates a continuum measurement set (MS) named calibrated_final_cont.ms and continuum images using the MS created in [[Imaging_Prep | Prepare the data for imaging]] section. Commands can be found in scriptForImaging_template.py available on [https://github.com/aakepley/ALMAImagingScript github]. | ||
#'''[[ | #'''[[Self-Calibration_Template | Self-Calibration]]''' This page provides a basic template for phase and amplitude self calibration of the continuum using the MS created in [[Image_Continuum | Image the Continuum]] section. Commands can be found in scriptForImaging_template.py available on [https://github.com/aakepley/ALMAImagingScript github]. | ||
#'''[[Image_Line | Spectral Line Imaging]]''' This page performs continuum subtraction and creates spectral line cubes using the MS created in [[Imaging_Prep | Prepare the data for imaging] section. Commands can be found in | #'''[[Image_Line | Spectral Line Imaging]]''' This page performs continuum subtraction and creates spectral line cubes using the MS created in [[Imaging_Prep | Prepare the data for imaging]] section. Commands can be found in scriptForImaging_template.py available on [https://github.com/aakepley/ALMAImagingScript github]. | ||
The NA imaging workflow used with these scripts is shown below. | The NA imaging workflow used with these scripts is shown below. | ||
[[Image:Imaging_WorkFlow.png|center|frame|1200px]] | [[Image:Imaging_WorkFlow.png|center|frame|1200px]] | ||
The older CASA 6.1 versions of these guides are available at the following links: | |||
#'''[[Imaging Prep CASA 6.5.4]]''' | |||
#'''[[Image Continuum CASA 6.5.4]]''' | |||
#'''[[Self-Calibration Template CASA 6.5.4]]''' | |||
#'''[[Image Line CASA 6.5.4]]''' | |||
==How to Use A CASA Guide== | ==How to Use A CASA Guide== | ||
See the [ | See the [https://casaguides.nrao.edu/index.php/CASA_Tips CASA Tips] page for tips on using CASA and ways CASA can be run. | ||
See [http://casaguides.nrao.edu/index.php?title=Extracting_scripts_from_these_tutorials Extracting Scripts From These Tutorials] for information on how to download the scripts from these tutorials. Note that the full scripts | See [http://casaguides.nrao.edu/index.php?title=Extracting_scripts_from_these_tutorials Extracting Scripts From These Tutorials] for information on how to download the scripts from these tutorials. Note that the full scripts can be downloaded via [https://github.com/aakepley/ALMAImagingScript github]. | ||
Within the guides: | Within the guides: | ||
Latest revision as of 16:40, 14 November 2024
About this Guide
This guide describes the template scripts used to manually image data for quality assurance by the North America ALMA Science Center (NAASC). This script is designed to guide the user through the decisions needed when imaging ALMA data. It should not be applied blindly; some editing of the scripts will be required in almost all cases. Not all sections of the template are relevant for all data sets. It can be adapted for use with partial pipeline processing of images. For example, you could do continuum subtraction in the pipeline, but use the scripts here as a template for manual imaging. See the ALMA Imaging Pipeline Reprocessing guide for information on pipeline processed images.
A current version of the script can be found on github. A description of how the script is used at the NAASC can be found in Kepley et al. Proceedings of the SPIE, 9910, 99100C .
More information on the ALMA Quality Assurance procedure and standards can be found on the ALMA Science Portal.
All example images were produced using ALMA Science Verification Data of TW Hya. More information along with the dataset can be obtained from the TW Hydra Band 7 Calibration and Imaging Guide.
Prepare for Imaging
Before you begin imaging, determine whether the dataset was manually or pipeline calibrated. The README file should state how the dataset was calibrated. If it does not, look in the script directory of the delivered data package. If you see a file named <ASDM UID>.ms.scriptForCalibration.py, the dataset was manually calibrated. If you see a filename PPR_*.xml, the dataset was pipeline calibrated.
If the data was calibrated prior to CASA 4.3, we recommend that you image the data in the same version of CASA that it was calibrated in because the weighting scheme for the data in CASA was in limbo. See Data Weights and Combination for more information. If the data was calibrated in CASA version 4.3 or later, imaging the data in a later version of CASA should work but we recommend using the same version of Casa used for calibration when imaging since Casa is not guaranteed to be backwards compatible. The README or CASA log file will indicate which CASA version was used to calibrate the data. Different versions of CASA can be obtained from Download CASA. Remember to check the operating system requirements before proceeding with the installation.
Instructions on restoring the calibrated measurement set should be found in the README of your delivered dataset. For most cases, this can be done by running CASA in the script directory and executing scriptForPI.py in CASA:
# in CASA
execfile(“scriptForPI.py”)
Attached Scripts
Two template scripts (scriptForImagingPrep_template.py and scriptForImaging_template.py) are available for this guide. These contain commands with minimal instructions and are intended to be used by those more familiar with imaging. A current version of the scripts can be found on github.
Obtaining Analysis Utilities
Some optional steps in this guide use the Analysis Utilities package. If you wish to use any of these tasks, follow the link above for download and installation instructions.
Background Information
This guide assumes that you are familiar with basic Python. If you need more information about python, see the official Python 3.13 tutorial
If you are new to interferometry or CASA, we recommend you follow the “First Look” Imaging Tutorials. The Synthesis Imaging Summer School Lectures provide a wealth of information on interferometry.
In addition, many frequently asked questions can be found in the ALMA Knowledgebase. If you have additional questions after reviewing this guide, please contact the ALMA Helpdesk.
Guide Contents
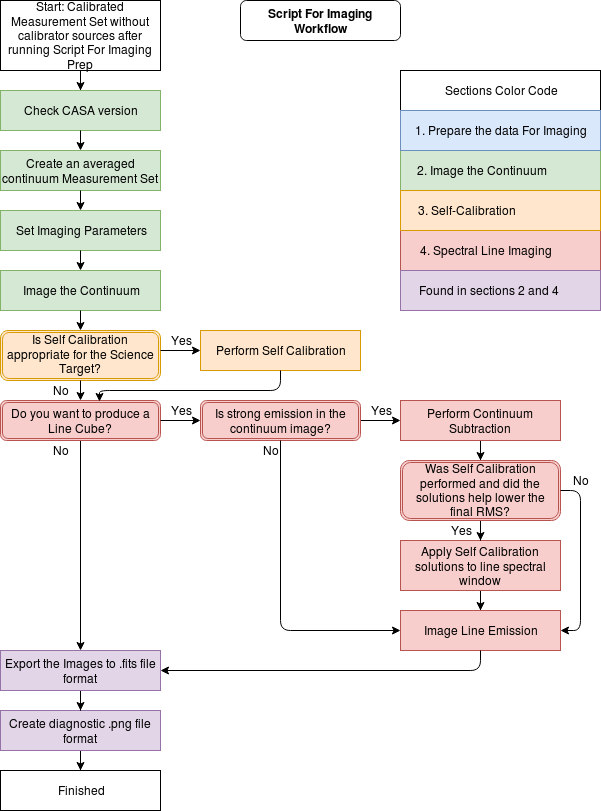
This guide has been split into 4 sections:
- Prepare the data for Imaging This page creates the calibrated_final.ms that will be used for imaging. Commands can be found in scriptForImagingPrep_template.py available on github.
- Image the Continuum This page creates a continuum measurement set (MS) named calibrated_final_cont.ms and continuum images using the MS created in Prepare the data for imaging section. Commands can be found in scriptForImaging_template.py available on github.
- Self-Calibration This page provides a basic template for phase and amplitude self calibration of the continuum using the MS created in Image the Continuum section. Commands can be found in scriptForImaging_template.py available on github.
- Spectral Line Imaging This page performs continuum subtraction and creates spectral line cubes using the MS created in Prepare the data for imaging section. Commands can be found in scriptForImaging_template.py available on github.
The NA imaging workflow used with these scripts is shown below.
The older CASA 6.1 versions of these guides are available at the following links:
- Imaging Prep CASA 6.5.4
- Image Continuum CASA 6.5.4
- Self-Calibration Template CASA 6.5.4
- Image Line CASA 6.5.4
How to Use A CASA Guide
See the CASA Tips page for tips on using CASA and ways CASA can be run.
See Extracting Scripts From These Tutorials for information on how to download the scripts from these tutorials. Note that the full scripts can be downloaded via github.
Within the guides:
# Regions of this color are CASA commands (or definitions) that need to be cut and
# pasted in sequence. Wait until one command is finished before pasting another.
# Tabs matter in python, make sure that commands that span more than one line and
# "for" loops keep their spacing. Sometimes (especially "for" loops) you may need to
# explicitly hit enter twice to get the command going.
Information in this color shows excerpts from the CASA Logger output
This color shows you background information about the data or other types of reference material