Calibrating an EVLA OSRO HI data set: Difference between revisions
| (50 intermediate revisions by the same user not shown) | |||
| Line 16: | Line 16: | ||
casapy # in your preferred shell | casapy # in your preferred shell | ||
</source> | </source> | ||
And import your SDM into CASA using [[importevla]]: | |||
</div> | </div> | ||
| Line 26: | Line 28: | ||
|} | |} | ||
[[Image:leo_importevla.png|800px|Importevla parameters.]] | |||
<div style="background-color: #dddddd;"> | |||
'''Tip:''' You'll notice that some parameters are colored differently than others. Standard parameters are simple black text (like '''ocorr_mode'''). Parameters which have been changed from their defaults are colored <span style="color:blue">blue</span> (e.g., '''vis'''). Parameters which are incorrectly set will turn <span style="color:red">red</span> (say, if you input a string and CASA expects an integer). Parameters which are <span style="background-color:darkgrey">back-shaded grey</span> (like '''singledish''') are expandable, and once they have been expanded (like '''applyflags'''), <span style="color:green">green</span> sub-parameters (like '''tbuff''') will appear. Note that if you alter an expandable parameter, a different menu of green sub-parameters will appear. | |||
</div> | |||
If you prefer to type it all one line (or use the script extractor), it will look like this: | |||
<source lang="python"> | <source lang="python"> | ||
importevla(asdm='leo2pt.55183.452640752315',vis='leotest.ms',corr_mode='co') | |||
asdm | |||
vis | |||
corr_mode | |||
</source> | </source> | ||
The archive spit out an SDM with the rather clumsy name 'leo2pt.55183.452640752315'. We're using [[importevla]] to create a measurement set out of it called '''vis=''' 'leotest.ms'. The measurement set is what we will use for the rest of the data reduction steps described below. After running [[importevla]], it will be a directory called 'leotest.ms/' in your working directory; You can check to see if it is there by typing 'ls'. You'll also want to set '''ocorr_mode=''' 'co', as the EVLA autocorrelation data is not generally useful. | |||
You might want a copy of your raw measurement set as a backup; You can just use <tt> cp -r leotest.ms leotestcp.ms'</tt> on the CASA or LINUX command line, or you can use [[split]]. | |||
[[Image:leo_split.png|800px|Split parameters.]] | |||
<source lang="python"> | |||
<source lang="python"> | |||
vis | split(vis='leotest.ms',outputvis='leotestcp.ms',datacolumn='data') | ||
outputvis | |||
datacolumn | |||
</source> | </source> | ||
It doesn't much matter now if you choose '''datacolumn=''' 'data' or 'corrected', as you have not calibrated the data yet, so the 'data' and 'corrected' columns will be identical. | |||
== Get Some Basic Information on The Data == | == Get Some Basic Information on The Data == | ||
Use [[listobs]] (roughly equivalent to LISTR in AIPS) to list information about the dataset's scans, correlator setup, and antenna positions. | Use [[listobs]] (roughly equivalent to LISTR in AIPS) to list information about the dataset's scans, correlator setup, and antenna positions. | ||
[[Image:leo_listobs.png|800px|Listobs parameters.]] | |||
<source lang="python"> | <source lang="python"> | ||
listobs(vis='leotest.ms') | |||
vis | |||
</source> | </source> | ||
[[Listobs]] will produce output that looks like this: | [[Listobs]] will produce output that looks like this: | ||
<pre> | <pre style="background-color: #fffacd;"> | ||
2010-02-11 18:11:34 INFO listobs ########################################## | 2010-02-11 18:11:34 INFO listobs ########################################## | ||
2010-02-11 18:11:34 INFO listobs ##### Begin Task: listobs ##### | 2010-02-11 18:11:34 INFO listobs ##### Begin Task: listobs ##### | ||
| Line 169: | Line 145: | ||
We'd like our reference antenna to be near the center of the array, and the easiest way to envision the array layout is with [[plotants]]. | We'd like our reference antenna to be near the center of the array, and the easiest way to envision the array layout is with [[plotants]]. | ||
< | <pre style="background-color: #fffacd;"> | ||
# plotants :: Plot the antenna distribution in the local reference frame: | # plotants :: Plot the antenna distribution in the local reference frame: | ||
vis = 'leotest.ms' # Name of input visibility file (MS) | vis = 'leotest.ms' # Name of input visibility file (MS) | ||
| Line 175: | Line 151: | ||
async = False # If true the taskname must be started using | async = False # If true the taskname must be started using | ||
# plotants(...) | # plotants(...) | ||
</ | </pre> | ||
[[Plotants]] will produce a plot that looks like this: | [[Plotants]] will produce a plot that looks like this: | ||
| Line 185: | Line 161: | ||
== First Round of Data Flagging == | == First Round of Data Flagging == | ||
Presently, no flags whatsoever are automatically applied to EVLA data. To apply online flags (for when the telescope is not on source or when the subreflector has not settled), you'll have to use the ''flagant.py'' script; see [[Applying_EVLA_on-line_flags | here]] for directions. | |||
After applying the online flags, check your calibrator data to see if the first few integrations of each scan are bad. If they are, flag them with [[flagdata]] in 'quack' mode. This is what the default [[flagdata]] parameters look like in our CASA window: | |||
[[Image:Flagdata_inp.jpg| | [[Image:Flagdata_inp.jpg|800px|Flagdata parameters.]] | ||
<div style="background-color: #dddddd;"> | <div style="background-color: #dddddd;"> | ||
'''Tip:''' You'll notice that some parameters are colored differently than others. Standard parameters (familiar from AIPS) are simple black text (like '''vis'''). Parameters which are shaded grey (like '''selectdata''') are expandable, and once they have been expanded (like '''mode'''), green sub-parameters (like '''autocorr''') will appear. Note that if you were to change '''mode''' from 'manualflag' to, say, 'quack', a different menu of green sub-parameters would appear. | '''Tip:''' You'll notice that some parameters are colored differently than others. Standard parameters (familiar from AIPS) are simple black text (like '''vis'''). Parameters which are shaded grey (like '''selectdata''') are expandable, and once they have been expanded (like '''mode'''), green sub-parameters (like '''autocorr''') will appear. Note that if you were to change '''mode''' from 'manualflag' to, say, 'quack', a different menu of green sub-parameters would appear. | ||
</div> | </div> | ||
To | To flag the first 10 seconds of each scan, set the [[flagdata]] parameters to: | ||
< | <pre style="background-color: #fffacd;"> | ||
# flagdata :: All purpose flagging task based on selections | # flagdata :: All purpose flagging task based on selections | ||
vis = 'leotest.ms' # Name of file to flag | vis = 'leotest.ms' # Name of file to flag | ||
| Line 211: | Line 189: | ||
selectdata = False # More data selection parameters (antenna, timerange etc) | selectdata = False # More data selection parameters (antenna, timerange etc) | ||
async = False # If true the taskname must be started using flagdata(...) | async = False # If true the taskname must be started using flagdata(...) | ||
</ | </pre> | ||
Currently, antenna shadowing is not included in the online flags, so you might have some shadowed antennas in your data. You can also use [[flagdata]] to edit these out, with '''mode=''' 'shadow'. | |||
Currently, online flags | <pre style="background-color: #fffacd;"> | ||
< | |||
# flagdata :: All purpose flagging task based on selections | # flagdata :: All purpose flagging task based on selections | ||
vis = 'leotest.ms' # Name of file to flag | vis = 'leotest.ms' # Name of file to flag | ||
| Line 225: | Line 202: | ||
selectdata = False # More data selection parameters (antenna, timerange etc) | selectdata = False # More data selection parameters (antenna, timerange etc) | ||
async = False # If true the taskname must be started using flagdata(...) | async = False # If true the taskname must be started using flagdata(...) | ||
</ | </pre> | ||
We conservatively use a diameter which is a bit bigger than the actual 25m diameter of the EVLA antennae. | We conservatively use a diameter which is a bit bigger than the actual 25m diameter of the EVLA antennae. | ||
Now is also the time to flag any obviously aberrant data | Finally, you might want to use [[flagdata]] to flag antennas with no receivers or other serious issues. Here is an example where we flag antenna ea13. | ||
<pre style="background-color: #fffacd;"> | |||
# flagdata :: All purpose flagging task based on selections | |||
vis = 'leotest.ms' # Name of file to flag | |||
flagbackup = True # Automatically back up the current flags? | |||
mode = 'manualflag' # Mode (manualflag,shadow,quack,summary,autoflag,rfi) | |||
autocorr = False # Flag autocorrelations | |||
unflag = False # Unflag the data specified | |||
clipexpr = '' # Expression to clip on | |||
clipminmax = [] # Range to use for clipping | |||
clipcolumn = 'DATA' # Data column to use for clipping | |||
clipoutside = True # Clip outside the range, or within it | |||
channelavg = False # Average over channels | |||
spw = '' # spectral-window/frequency/channel | |||
field = '' # Field names or field index numbers: ''==>all, field='0~2,3C286' | |||
selectdata = True # More data selection parameters (antenna, timerange etc) | |||
antenna = 'ea13' # antenna/baselines: ''==>all, antenna = '3,VA04' | |||
timerange = '' # time range: ''==>all, timerange='09:14:0~09:54:0' | |||
correlation = '' # Select data based on correlation | |||
scan = '' # scan numbers: ''==>all | |||
feed = '' # multi-feed numbers: Not yet implemented | |||
array = '' # (sub)array numbers: ''==>all | |||
uvrange = '' # uv range: ''==>all; uvrange = '0~100klambda', default | |||
# units=meters | |||
async = False # If true the taskname must be started using flagdata(...) | |||
</pre> | |||
Now is also the time to flag any obviously aberrant data. You might choose to flag with [[viewer]], which displays a raster image of your uv data and is similar to SPFLG and TVFLG in AIPS [[Data_flagging_with_viewer | (click here for an introduction to viewer)]]. Alternatively, you can flag using a 2-dimensional interactive plot with [[plotms]] [[Data_flagging_with_plotms | (click here for an introduction to plotms)]]. The plotting capabilities can be likened to UVPLT in AIPS, and data points can be flagged (like WIPER in AIPS). | |||
== Calibration == | == Calibration == | ||
| Line 234: | Line 240: | ||
=== Set the Flux Scale === | === Set the Flux Scale === | ||
Use the task [[setjy]] to find the flux density of the flux calibrator (as in AIPS). In this case, the flux calibrator is 1331+305. | Use the task [[setjy]] to find the flux density of the flux calibrator (as in AIPS). In this case, the flux calibrator is 1331+305. | ||
< | <pre style="background-color: #fffacd;"> | ||
# setjy :: Fills the model column with the visibility of a calibrator | # setjy :: Fills the model column with the visibility of a calibrator | ||
vis = 'leotest.ms' # Name of input visibility file | vis = 'leotest.ms' # Name of input visibility file | ||
| Line 243: | Line 249: | ||
standard = 'Perley-Taylor 99' # Flux density standard | standard = 'Perley-Taylor 99' # Flux density standard | ||
async = False # If true the taskname must be started using setjy(...) | async = False # If true the taskname must be started using setjy(...) | ||
</ | </pre> | ||
In most cases, you should be wary that the flux calibrator could be resolved, and you should use a model (set by the '''modimage''' parameter above). CASA comes pre-packed with model images, but finding them might be a bit tricky. Here are some probable paths (see the [http://casa.nrao.edu/Doc/Cookbook/casa_cookbook.pdf CASA Cookbook] for more detail): | In most cases, you should be wary that the flux calibrator could be resolved, and you should use a model (set by the '''modimage''' parameter above). CASA comes pre-packed with model images, but finding them might be a bit tricky. Here are some probable paths (see the [http://casa.nrao.edu/Doc/Cookbook/casa_cookbook.pdf CASA Cookbook] for more detail): | ||
* Red Hat Linux RPMs (RHE4, Fedora 6): /usr/lib/casapy/data/nrao/VLA/CalModels | * Red Hat Linux RPMs (RHE4, Fedora 6): /usr/lib/casapy/data/nrao/VLA/CalModels | ||
* MAC OSX .dmg: /opt/casa/data/nrao/VLA/CalModels | * MAC OSX .dmg: /opt/casa/data/nrao/VLA/CalModels | ||
* NRAO-AOC stable: /home/casa/data/nrao/VLA/CalModels | * NRAO-AOC stable: /home/casa/data/nrao/VLA/CalModels | ||
[[Setjy]] will copy this model to the 'model' data column of your measurement set, and use it in subsequent calibrations of '1331+305'. | |||
You should also note that [[setjy]] expects your flux calibrator to be named in certain conventions (See Table 4.1 of the [http://casa.nrao.edu/Doc/Cookbook/casa_cookbook.pdf CASA Cookbook] for acceptable names). If your observations don't follow these conventions, you'll probably have to [[Renaming_a_Field | change the flux calibrator's field name]]. | You should also note that [[setjy]] expects your flux calibrator to be named in certain conventions (See Table 4.1 of the [http://casa.nrao.edu/Doc/Cookbook/casa_cookbook.pdf CASA Cookbook] for acceptable names). If your observations don't follow these conventions, you'll probably have to [[Renaming_a_Field | change the flux calibrator's field name]]. | ||
[[Setjy]] should output something that looks like this: | [[Setjy]] should output something that looks like this: | ||
<pre> | <pre style="background-color: #ffe4b5;"> | ||
2010-02-11 23:57:53 INFO setjy ########################################## | 2010-02-11 23:57:53 INFO setjy ########################################## | ||
2010-02-11 23:57:53 INFO setjy ##### Begin Task: setjy ##### | 2010-02-11 23:57:53 INFO setjy ##### Begin Task: setjy ##### | ||
| Line 273: | Line 281: | ||
To optimize our bandpass calibration, we'll first need to do a preliminary phase calibration on 1331+305 with [[gaincal]] (analogous to CALIB in AIPS). | To optimize our bandpass calibration, we'll first need to do a preliminary phase calibration on 1331+305 with [[gaincal]] (analogous to CALIB in AIPS). | ||
< | <pre style="background-color: #fffacd;"> | ||
# gaincal :: Determine temporal gains from calibrator observations | # gaincal :: Determine temporal gains from calibrator observations | ||
vis = 'leotest.ms' # Nome of input visibility file | vis = 'leotest.ms' # Nome of input visibility file | ||
| Line 299: | Line 307: | ||
parang = False # Apply parallactic angle correction on the fly | parang = False # Apply parallactic angle correction on the fly | ||
async = False # If true the taskname must be started using gaincal(...) | async = False # If true the taskname must be started using gaincal(...) | ||
</ | </pre> | ||
You'll want to find an intuitive naming convention for your calibration tables; here, we output the gain calibration to a table called 'leotest_0.gcal'. The above parameters calibrate only the phases, not amplitudes ('''calmode=''' 'p') for the flux calibrator ('''field=''' '1331+305')'. We use a narrow channel range in the center of the bandpass ('''spw=''' '*:118~137'), chosen so that the bandpass does not vary much across this range. The combination of parameters '''solint=''' 'inf' and '''combine=''' ' ' tell [[gaincal]] to time-integrate across an entire calibrator scan in finding a solution. We have assigned a source model to the flux calibrator with [[setjy]], so we do not need to set a '''uvrange'''. Don't forget to set the reference antenna ('''refant=''' 'ea25'). | You'll want to find an intuitive naming convention for your calibration tables; here, we output the gain calibration to a table called 'leotest_0.gcal'. The above parameters calibrate only the phases, not amplitudes ('''calmode=''' 'p') for the flux calibrator ('''field=''' '1331+305')'. We use a narrow channel range in the center of the bandpass ('''spw=''' '*:118~137'; see [[Selecting_Spectral_Windows_and_Channels|this description]] for more detail on selecting spectral windows and channels), chosen so that the bandpass does not vary much across this range. The combination of parameters '''solint=''' 'inf' and '''combine=''' ' ' tell [[gaincal]] to time-integrate across an entire calibrator scan in finding a solution. We have assigned a source model to the flux calibrator with [[setjy]], so we do not need to set a '''uvrange'''. Don't forget to set the reference antenna ('''refant=''' 'ea25'). | ||
The calibration tables will be outputted as a directory in your present working directory. If you type 'ls' on the CASA command line, you should see a directory named for your '''caltable'''. | The calibration tables will be outputted as a directory in your present working directory. If you type 'ls' on the CASA command line, you should see a directory named for your '''caltable'''. | ||
| Line 308: | Line 316: | ||
Check that the phase calibration solutions look reasonable with [[plotcal]] (which, in this case, is similar to SNPLT in AIPS). | Check that the phase calibration solutions look reasonable with [[plotcal]] (which, in this case, is similar to SNPLT in AIPS). | ||
< | <pre style="background-color: #fffacd;"> | ||
# plotcal :: An all-purpose plotter for calibration results | # plotcal :: An all-purpose plotter for calibration results | ||
caltable = 'leotest_0.gcal' # Name of input calibration table | caltable = 'leotest_0.gcal' # Name of input calibration table | ||
| Line 331: | Line 339: | ||
figfile = '' # ''= no plot hardcopy, otherwise supply name | figfile = '' # ''= no plot hardcopy, otherwise supply name | ||
async = False # If true the taskname must be started using plotcal(...) | async = False # If true the taskname must be started using plotcal(...) | ||
</ | </pre> | ||
[[File:Plotcal_gtable0.jpg | thumb | A phase vs. time plot from [[plotcal]].]] | [[File:Plotcal_gtable0.jpg | thumb | A phase vs. time plot from [[plotcal]].]] | ||
| Line 338: | Line 346: | ||
Now, we're ready to solve for bandpass solutions with [[bandpass]]: | Now, we're ready to solve for bandpass solutions with [[bandpass]]: | ||
< | <pre style="background-color: #fffacd;"> | ||
# bandpass :: Calculates a bandpass calibration solution | # bandpass :: Calculates a bandpass calibration solution | ||
vis = 'leotest.ms' # Nome of input visibility file | vis = 'leotest.ms' # Nome of input visibility file | ||
| Line 362: | Line 370: | ||
parang = False # Apply parallactic angle correction | parang = False # Apply parallactic angle correction | ||
async = False # If true the taskname must be started using bandpass(...) | async = False # If true the taskname must be started using bandpass(...) | ||
</ | </pre> | ||
Important parameters to consider are: | Important parameters to consider are: | ||
* '''caltable''': the name of the bandpass calibration table that will be generated | * '''caltable''': the name of the bandpass calibration table that will be generated | ||
| Line 373: | Line 381: | ||
After running [[bandpass]] and getting some helpful output in the CASA logger, we can also plot up our bandpass solutions with [[plotcal]] (which, in this case, can be likened to POSSM in AIPS with APARM(8)=2). [[Plotcal]] parameters that look like this: | After running [[bandpass]] and getting some helpful output in the CASA logger, we can also plot up our bandpass solutions with [[plotcal]] (which, in this case, can be likened to POSSM in AIPS with APARM(8)=2). [[Plotcal]] parameters that look like this: | ||
< | <pre style="background-color: #fffacd;"> | ||
# plotcal :: An all-purpose plotter for calibration results | # plotcal :: An all-purpose plotter for calibration results | ||
caltable = 'leotest_1.bcal' # Name of input calibration table | caltable = 'leotest_1.bcal' # Name of input calibration table | ||
| Line 396: | Line 404: | ||
figfile = '' # ''= no plot hardcopy, otherwise supply name | figfile = '' # ''= no plot hardcopy, otherwise supply name | ||
async = False # If true the taskname must be started using plotcal(...) | async = False # If true the taskname must be started using plotcal(...) | ||
</ | </pre> | ||
[[File:Bandpass.jpg | thumb | A bandpass plot from [[plotcal]].]] | [[File:Bandpass.jpg | thumb | A bandpass plot from [[plotcal]].]] | ||
| Line 411: | Line 419: | ||
=== Calibrate the Gains === | === Calibrate the Gains === | ||
Now it's time for the amplitude and phase calibration with [[gaincal]]. If you are going to use the same '''uvrange''' for all calibrator sources, then you can run [[gaincal]] simultaneously on them all. Check with the [http://www.vla.nrao.edu/astro/calib/manual/csource.html calibrator manual] to see the acceptable '''uvrange''' for each of your calibrators (if you have assigned a source model to your flux calibrator with [[setjy]], then for that source you can ignore the suggested '''uvrange''' in the calibrator manual and use the full range). Here our phase calibrator (J1042+1203) has a suggested '''uvrange''' of 0--5 kilolambda, so we are going to run [[gaincal]] separately for the flux calibrator and the phase calibrator. | Now it's time for the amplitude and phase calibration with [[gaincal]]. If you are going to use the same '''uvrange''' for all calibrator sources, then you can run [[gaincal]] simultaneously on them all. Check with the [http://www.vla.nrao.edu/astro/calib/manual/csource.html calibrator manual] to see the acceptable '''uvrange''' for each of your calibrators (if you have assigned a source model to your flux calibrator with [[setjy]], then for that source you can ignore the suggested '''uvrange''' in the calibrator manual and use the full range). Here our phase calibrator (J1042+1203) has a suggested '''uvrange''' of 0--5 kilolambda, so we are going to run [[gaincal]] separately for the flux calibrator and the phase calibrator. | ||
< | <pre style="background-color: #fffacd;"> | ||
# gaincal :: Determine temporal gains from calibrator observations | # gaincal :: Determine temporal gains from calibrator observations | ||
vis = 'leotest.ms' # Nome of input visibility file | vis = 'leotest.ms' # Nome of input visibility file | ||
| Line 436: | Line 444: | ||
parang = False # Apply parallactic angle correction on the fly | parang = False # Apply parallactic angle correction on the fly | ||
async = False # If true the taskname must be started using gaincal(...) | async = False # If true the taskname must be started using gaincal(...) | ||
</ | </pre> | ||
The above parameters calibrate the amplitudes and phases ('''calmode=''' 'ap') for the flux calibrator ('''field=''' '1331+305') and outputs the gain calibration to a table called 'leotest_1.gcal'. We only use channels 25--250 to calibrate ('''spw=''' '*:25~250'), chosen by visual inspection of the bandpass solutions with [[plotcal]]. We use one solution time interval per scan ('''solint=''' 'inf' and '''combine=''' ' '). In order for flux bootstrapping to work properly in [[Reducing_an_EVLA_OSRO_HI_data_set#Apply_the_Flux_Scale | the next calibration step]], we want to refrain from normalizing the solutions ('''solnorm=''' False). Don't forget to set the reference antenna ('''refant=''' 'ea25') and apply the bandpass calibration ('''gaintable=''' 'leotest_1.bcal'). | The above parameters calibrate the amplitudes and phases ('''calmode=''' 'ap') for the flux calibrator ('''field=''' '1331+305') and outputs the gain calibration to a table called 'leotest_1.gcal'. We only use channels 25--250 to calibrate ('''spw=''' '*:25~250'), chosen by visual inspection of the bandpass solutions with [[plotcal]]. Gaincal will apply the bandpass calibration ('leotest_1.bcal') to these channels, average them together, and then find gain solutions. We use one solution time interval per scan ('''solint=''' 'inf' and '''combine=''' ' '). In order for flux bootstrapping to work properly in [[Reducing_an_EVLA_OSRO_HI_data_set#Apply_the_Flux_Scale | the next calibration step]], we want to refrain from normalizing the solutions ('''solnorm=''' False). Don't forget to set the reference antenna ('''refant=''' 'ea25') and apply the bandpass calibration ('''gaintable=''' 'leotest_1.bcal'). | ||
Next, we will calibrate the phase calibrator ('''field=''' 'J1042+1203') and set a limited '''uvrange''' ('''uvrange=''' '0~5klambda'): | Next, we will calibrate the phase calibrator ('''field=''' 'J1042+1203') and set a limited '''uvrange''' ('''uvrange=''' '0~5klambda'): | ||
< | <pre style="background-color: #fffacd;"> | ||
# gaincal :: Determine temporal gains from calibrator observations | # gaincal :: Determine temporal gains from calibrator observations | ||
vis = 'leotest.ms' # Nome of input visibility file | vis = 'leotest.ms' # Nome of input visibility file | ||
| Line 470: | Line 478: | ||
parang = False # Apply parallactic angle correction on the fly | parang = False # Apply parallactic angle correction on the fly | ||
async = False # If true the taskname must be started using gaincal(...) | async = False # If true the taskname must be started using gaincal(...) | ||
</ | </pre> | ||
[[File:Plotcal_gtable.jpg | thumb | Plot of amplitude vs. time from [[plotcal]].]] | [[File:Plotcal_gtable.jpg | thumb | Plot of amplitude vs. time from [[plotcal]].]] | ||
| Line 481: | Line 489: | ||
Next, we'll bootstrap the flux scale that was found by running [[setjy]] onto our phase calibrator with the CASA task [[fluxscale]] (synonymous with GETJY in AIPS). | Next, we'll bootstrap the flux scale that was found by running [[setjy]] onto our phase calibrator with the CASA task [[fluxscale]] (synonymous with GETJY in AIPS). | ||
< | <pre style="background-color: #fffacd;"> | ||
# fluxscale :: Bootstrap the flux density scale from standard calibrators | # fluxscale :: Bootstrap the flux density scale from standard calibrators | ||
vis = 'leotest.ms' # Name of input visibility file (MS) | vis = 'leotest.ms' # Name of input visibility file (MS) | ||
| Line 491: | Line 499: | ||
refspwmap = [-1] # Scale across spectral window boundaries. See help fluxscale | refspwmap = [-1] # Scale across spectral window boundaries. See help fluxscale | ||
async = False # If true the taskname must be started using fluxscale(...) | async = False # If true the taskname must be started using fluxscale(...) | ||
</ | </pre> | ||
We get output like this: | We get output like this: | ||
<pre> | <pre style="background-color: #ffe4b5;"> | ||
2010-02-14 21:35:04 INFO fluxscale ########################################## | 2010-02-14 21:35:04 INFO fluxscale ########################################## | ||
2010-02-14 21:35:04 INFO fluxscale ##### Begin Task: fluxscale ##### | 2010-02-14 21:35:04 INFO fluxscale ##### Begin Task: fluxscale ##### | ||
| Line 508: | Line 516: | ||
2010-02-14 21:35:04 INFO fluxscale ##### End Task: fluxscale ##### | 2010-02-14 21:35:04 INFO fluxscale ##### End Task: fluxscale ##### | ||
</pre> | </pre> | ||
which tells us that our phase calibrator has a flux density of 3.1 Jy. Here, it is good to be on guard for flux densities which are suspiciously close to 1 Jy or errors in the derived flux density which are very large. Note that, unlike in AIPS, [[fluxscale]] writes a new calibration table, and this is what we will want to apply to our data. | which tells us that our phase calibrator has a flux density of 3.1 Jy. Here, it is good to be on guard for flux densities which are suspiciously close to 1 Jy or errors in the derived flux density which are very large. Note that, unlike in AIPS, [[fluxscale]] writes a new calibration table, and this is what we will want to apply to our data. The output table 'leotest_1.fluxscale' does not only include information about the flux density scale, but also includes the full solution of gains, copied over from 'leotest_1.gcal'---so it supersedes 'leotest_1.gcal'. | ||
=== Apply the Calibration === | === Apply the Calibration === | ||
Now, we're happy with our calibration and ready to apply it to the data with [[applycal]]. | Now, we're happy with our calibration and ready to apply it to the data with [[applycal]]. You'll want to give [[applycal]] ''all'' relevant calibration tables! Here we are going to calibrate bandpasses, gains, and flux densities. | ||
< | |||
[[Applycal]] might be compared with CLCAL in AIPS, except that instead of writing a new calibration (CL) table, [[applycal]] will write a new calibrated column of data in your '.ms' file. It also differs from CLCAL in that we will use [[applycal]] to apply ''all'' calibrations, not only gain/flux density solutions. After we run [[applycal]], we will still be able to access the raw data (called 'data' by many tasks), but there will also be calibrated data (called 'corrected' by many tasks). | |||
<pre style="background-color: #fffacd;"> | |||
# applycal :: Apply calibrations solutions(s) to data | # applycal :: Apply calibrations solutions(s) to data | ||
vis = 'leotest.ms' # Nome of input visibility file | vis = 'leotest.ms' # Nome of input visibility file | ||
| Line 527: | Line 537: | ||
calwt = True # Calibrate data weights from all relevant calibrations | calwt = True # Calibrate data weights from all relevant calibrations | ||
async = False # If true the taskname must be started using applycal(...) | async = False # If true the taskname must be started using applycal(...) | ||
</ | </pre> | ||
The above settings for [[applycal]] will calibrate all sources, linearly interpolating between [[gaincal]] solutions. It will apply the flux, gain, ''and'' bandpass calibration ('''gaintable=''' ['leotest_1.bcal', 'leotest_1.fluxscale']). | The above settings for [[applycal]] will calibrate all sources, linearly interpolating between [[gaincal]] solutions. It will apply the flux, gain, ''and'' bandpass calibration ('''gaintable=''' ['leotest_1.bcal', 'leotest_1.fluxscale']). | ||
| Line 547: | Line 557: | ||
== Split Off the Science Targets == | == Split Off the Science Targets == | ||
Finally, we'll want to split off our calibrated science targets from the multi-source measurement set, so that we can image them! Use [[split]], | Finally, we'll want to split off our calibrated science targets from the multi-source measurement set, so that we can image them! Use [[split]], as you would in AIPS (However, note that [[split]] in CASA can not apply calibration as AIPS SPLIT does. You have to first apply the calibration with [[applycal]], the specify '''datacolumn=''' 'corrected'). | ||
< | <pre style="background-color: #fffacd;"> | ||
# split :: Create a visibility subset from an existing visibility set | # split :: Create a visibility subset from an existing visibility set | ||
vis = 'leotest.ms' # Name of input measurement set | vis = 'leotest.ms' # Name of input measurement set | ||
| Line 563: | Line 573: | ||
uvrange = '' # select data based on uv distance range | uvrange = '' # select data based on uv distance range | ||
async = False # If true the taskname must be started using split(...) | async = False # If true the taskname must be started using split(...) | ||
</ | </pre> | ||
The above will split off the calibrated data ('''datacolumn=''' 'corrected') for the first Leo field ('''field=''' 'Leo-1'). We'll want to repeat the above for the second Leo pointing, 'Leo-2'. | The above will split off the calibrated data ('''datacolumn=''' 'corrected') for the first Leo field ('''field=''' 'Leo-1'). We'll want to repeat the above for the second Leo pointing, 'Leo-2'. | ||
Now is also the time to note that all EVLA OSRO datasets are currently observed with 1 second integrations. This is likely to be more time resolution than you need for imaging, and your data set is probably quite large, making it time-consuming to reduce. Therefore, you might consider time averaging by increasing the '''timebin''' parameter from '0s' to your desired value). See [http://science.nrao.edu/evla/proposing/evlaoss/evlaoss-20100121/node21.html the EVLA Observational Status Summary] for advice on time averaging. | |||
Now we are ready to flag these science targets and image them! Continue on to [[Imaging_an_EVLA_OSRO_HI_data_set | this imaging tutorial]] for the next installment.... | Now we are ready to flag these science targets and image them! Continue on to [[Imaging_an_EVLA_OSRO_HI_data_set | this imaging tutorial]] for the next installment.... | ||
Latest revision as of 21:21, 4 May 2010
Overview
This article describes the calibration of an HI dataset collected on the Leo Ring with WIDAR0. This page will be updated when "real" OSRO data become available, but the below steps should be applicable to current EVLA data. Also see the Imaging an EVLA OSRO HI data set tutorial for the latter steps in the data reduction. We also note that this tutorial is written assuming you are reducing your data interactively, step-by-step; sorry, there are no python scripts to extract below with the script extractor.
Importing Data
|
To download your data, first find it in the NRAO archive. You'll have two options for file formats: either a science data model (SDM) or a measurement set (CASA MS). We recommend downloading the SDM, as this best ensures that your data will be compatible with the version of CASA you are using. You can also instruct the NRAO archive to tar the SDM. The figure to the right shows our selections in the archive for downloading the Leo data. Wait for an email from the NRAO archive with directions about downloading your data set, and then transfer the SDM to the directory that you will run CASA from. Start CASA in that directory. casapy # in your preferred shell
And import your SDM into CASA using importevla: |
Tip: You'll notice that some parameters are colored differently than others. Standard parameters are simple black text (like ocorr_mode). Parameters which have been changed from their defaults are colored blue (e.g., vis). Parameters which are incorrectly set will turn red (say, if you input a string and CASA expects an integer). Parameters which are back-shaded grey (like singledish) are expandable, and once they have been expanded (like applyflags), green sub-parameters (like tbuff) will appear. Note that if you alter an expandable parameter, a different menu of green sub-parameters will appear.
If you prefer to type it all one line (or use the script extractor), it will look like this:
importevla(asdm='leo2pt.55183.452640752315',vis='leotest.ms',corr_mode='co')
The archive spit out an SDM with the rather clumsy name 'leo2pt.55183.452640752315'. We're using importevla to create a measurement set out of it called vis= 'leotest.ms'. The measurement set is what we will use for the rest of the data reduction steps described below. After running importevla, it will be a directory called 'leotest.ms/' in your working directory; You can check to see if it is there by typing 'ls'. You'll also want to set ocorr_mode= 'co', as the EVLA autocorrelation data is not generally useful.
You might want a copy of your raw measurement set as a backup; You can just use cp -r leotest.ms leotestcp.ms' on the CASA or LINUX command line, or you can use split.
split(vis='leotest.ms',outputvis='leotestcp.ms',datacolumn='data')
It doesn't much matter now if you choose datacolumn= 'data' or 'corrected', as you have not calibrated the data yet, so the 'data' and 'corrected' columns will be identical.
Get Some Basic Information on The Data
Use listobs (roughly equivalent to LISTR in AIPS) to list information about the dataset's scans, correlator setup, and antenna positions.
listobs(vis='leotest.ms')
Listobs will produce output that looks like this:
2010-02-11 18:11:34 INFO listobs ########################################## 2010-02-11 18:11:34 INFO listobs ##### Begin Task: listobs ##### 2010-02-11 18:11:34 INFO listobs::::casa 2010-02-11 18:11:34 INFO listobs ================================================================================ 2010-02-11 18:11:34 INFO listobs MeasurementSet Name: /export/home/rso-lchomiuk/test/Leo/leotest.ms MS Version 2 2010-02-11 18:11:34 INFO listobs ================================================================================ 2010-02-11 18:11:34 INFO listobs Observer: leo2pt Project: T.B.D. 2010-02-11 18:11:34 INFO listobs Observation: VLA 2010-02-11 18:11:34 INFO listobs Data records: 70290 Total integration time = 11264.5 seconds 2010-02-11 18:11:34 INFO listobs Observed from 18-Dec-2009/10:52:17.0 to 18-Dec-2009/14:00:01.5 (UTC) 2010-02-11 18:11:35 INFO listobs::ms::summary 2010-02-11 18:11:35 INFO listobs ObservationID = 0 ArrayID = 0 2010-02-11 18:11:35 INFO listobs Date Timerange (UTC) Scan FldId FieldName nVis Int(s) SpwIds 2010-02-11 18:11:35 INFO listobs 18-Dec-2009/10:52:17.0 - 10:53:45.0 1 0 J1042+1203 660 9.6 [0] 2010-02-11 18:11:35 INFO listobs 10:54:14.0 - 10:56:51.5 2 1 J1042+1203 1122 9.71 [0] 2010-02-11 18:11:35 INFO listobs 10:57:20.0 - 11:06:57.5 3 2 Leo-1 3894 9.92 [0] 2010-02-11 18:11:35 INFO listobs 11:07:25.0 - 11:17:03.0 4 3 Leo-2 3894 9.93 [0] 2010-02-11 18:11:35 INFO listobs 11:17:32.0 - 11:22:10.0 5 2 Leo-1 1914 9.86 [0] 2010-02-11 18:11:35 INFO listobs 11:22:39.0 - 11:27:16.5 6 3 Leo-2 1914 9.83 [0] 2010-02-11 18:11:35 INFO listobs 11:27:45.0 - 11:30:23.5 7 1 J1042+1203 1122 9.82 [0] 2010-02-11 18:11:35 INFO listobs 11:30:52.0 - 11:40:30.0 8 2 Leo-1 3894 9.93 [0] 2010-02-11 18:11:35 INFO listobs 11:40:59.0 - 11:50:36.5 9 3 Leo-2 3894 9.92 [0] 2010-02-11 18:11:35 INFO listobs 11:51:05.0 - 11:55:42.5 10 2 Leo-1 1914 9.83 [0] 2010-02-11 18:11:35 INFO listobs 11:56:11.0 - 12:00:49.0 11 3 Leo-2 1914 9.86 [0] 2010-02-11 18:11:35 INFO listobs 12:01:18.0 - 12:03:56.0 12 1 J1042+1203 1122 9.76 [0] 2010-02-11 18:11:35 INFO listobs 12:04:25.0 - 12:14:03.0 13 2 Leo-1 3894 9.93 [0] 2010-02-11 18:11:35 INFO listobs 12:14:32.0 - 12:24:09.0 14 3 Leo-2 3894 9.9 [0] 2010-02-11 18:11:35 INFO listobs 12:24:37.0 - 12:29:15.0 15 2 Leo-1 1914 9.86 [0] 2010-02-11 18:11:35 INFO listobs 12:29:44.0 - 12:34:21.5 16 3 Leo-2 1914 9.83 [0] 2010-02-11 18:11:35 INFO listobs 12:34:49.0 - 12:37:28.0 17 1 J1042+1203 1122 9.88 [0] 2010-02-11 18:11:35 INFO listobs 12:37:57.0 - 12:47:35.0 18 2 Leo-1 3894 9.93 [0] 2010-02-11 18:11:35 INFO listobs 12:48:04.0 - 12:57:41.5 19 3 Leo-2 3894 9.92 [0] 2010-02-11 18:11:35 INFO listobs 12:58:10.0 - 13:02:48.0 20 2 Leo-1 1914 9.86 [0] 2010-02-11 18:11:35 INFO listobs 13:03:17.0 - 13:07:54.5 21 3 Leo-2 1914 9.83 [0] 2010-02-11 18:11:35 INFO listobs 13:08:23.0 - 13:11:01.5 22 1 J1042+1203 1122 9.82 [0] 2010-02-11 18:11:35 INFO listobs 13:11:31.0 - 13:21:08.5 23 2 Leo-1 3894 9.92 [0] 2010-02-11 18:11:35 INFO listobs 13:21:37.0 - 13:31:14.5 24 3 Leo-2 3894 9.92 [0] 2010-02-11 18:11:35 INFO listobs 13:31:43.0 - 13:36:21.0 25 2 Leo-1 1914 9.86 [0] 2010-02-11 18:11:35 INFO listobs 13:36:50.0 - 13:41:27.5 26 3 Leo-2 1914 9.83 [0] 2010-02-11 18:11:35 INFO listobs 13:41:56.0 - 13:44:34.5 27 1 J1042+1203 1122 9.82 [0] 2010-02-11 18:11:35 INFO listobs 13:48:02.0 - 14:00:01.5 28 4 1331+305 4818 9.99 [0] 2010-02-11 18:11:35 INFO listobs (nVis = Total number of time/baseline visibilities per scan) 2010-02-11 18:11:35 INFO listobs Fields: 5 2010-02-11 18:11:35 INFO listobs ID Code Name RA Decl Epoch SrcId nVis 2010-02-11 18:11:35 INFO listobs 0 NONE J1042+1203 10:42:44.6052 +12.03.31.2641 J2000 0 660 2010-02-11 18:11:35 INFO listobs 1 D J1042+1203 10:42:44.6052 +12.03.31.2641 J2000 1 6732 2010-02-11 18:11:35 INFO listobs 2 NONE Leo-1 10:47:22.0000 +12.16.38.0000 J2000 2 29040 2010-02-11 18:11:35 INFO listobs 3 NONE Leo-2 10:46:45.0000 +11.50.38.0000 J2000 3 29040 2010-02-11 18:11:35 INFO listobs 4 K 1331+305 13:31:08.2880 +30.30.32.9589 J2000 4 4818 2010-02-11 18:11:35 INFO listobs (nVis = Total number of time/baseline visibilities per field) 2010-02-11 18:11:35 INFO listobs Spectral Windows: (1 unique spectral windows and 1 unique polarization setups) 2010-02-11 18:11:35 INFO listobs SpwID #Chans Frame Ch1(MHz) ChanWid(kHz)TotBW(kHz) Ref(MHz) Corrs 2010-02-11 18:11:35 INFO listobs 0 256 TOPO 1415.3756 7.8125 2000 1415.3756 RR LL 2010-02-11 18:11:35 INFO listobs Sources: 6 2010-02-11 18:11:35 INFO listobs ID Name SpwId RestFreq(MHz) SysVel(km/s) 2010-02-11 18:11:35 INFO listobs 0 J1042+1203 0 - - 2010-02-11 18:11:35 INFO listobs 1 J1042+1203 0 - - 2010-02-11 18:11:35 INFO listobs 2 Leo-1 0 - - 2010-02-11 18:11:35 INFO listobs 3 Leo-2 0 - - 2010-02-11 18:11:35 INFO listobs 4 J1331+3030 0 - - 2010-02-11 18:11:35 INFO listobs 5 J1331+3030 0 - - 2010-02-11 18:11:35 INFO listobs Antennas: 12: 2010-02-11 18:11:35 INFO listobs ID Name Station Diam. Long. Lat. 2010-02-11 18:11:35 INFO listobs 0 ea02 E02 25.0 m -107.37.04.4 +33.54.01.1 2010-02-11 18:11:35 INFO listobs 1 ea03 E09 25.0 m -107.36.45.1 +33.53.53.6 2010-02-11 18:11:35 INFO listobs 2 ea04 W01 25.0 m -107.37.05.9 +33.54.00.5 2010-02-11 18:11:35 INFO listobs 3 ea05 W08 25.0 m -107.37.21.6 +33.53.53.0 2010-02-11 18:11:35 INFO listobs 4 ea08 N01 25.0 m -107.37.06.0 +33.54.01.8 2010-02-11 18:11:35 INFO listobs 5 ea09 E06 25.0 m -107.36.55.6 +33.53.57.7 2010-02-11 18:11:35 INFO listobs 6 ea15 W06 25.0 m -107.37.15.6 +33.53.56.4 2010-02-11 18:11:35 INFO listobs 7 ea19 W04 25.0 m -107.37.10.8 +33.53.59.1 2010-02-11 18:11:35 INFO listobs 8 ea24 W05 25.0 m -107.37.13.0 +33.53.57.8 2010-02-11 18:11:35 INFO listobs 9 ea25 N02 25.0 m -107.37.06.2 +33.54.03.5 2010-02-11 18:11:35 INFO listobs 10 ea27 E03 25.0 m -107.37.02.8 +33.54.00.5 2010-02-11 18:11:35 INFO listobs 11 ea28 N08 25.0 m -107.37.07.5 +33.54.15.8 2010-02-11 18:11:35 INFO listobs::::casa 2010-02-11 18:11:35 INFO listobs ##### End Task: listobs ##### 2010-02-11 18:11:35 INFO listobs ##########################################
Some things to note here are: The phase calibrator is called J1042+1203, the flux calibrator is 1331+305, and there are two science pointings, Leo-1 and Leo-2. The correlator is set up in such a way to have one spectral window (or IF). This window is sampled with 256 channels each 7.8 kHz wide. There are no cross-polarization data (note that 'Corrs' is set to only 'RR LL').
Choose a Reference Antenna
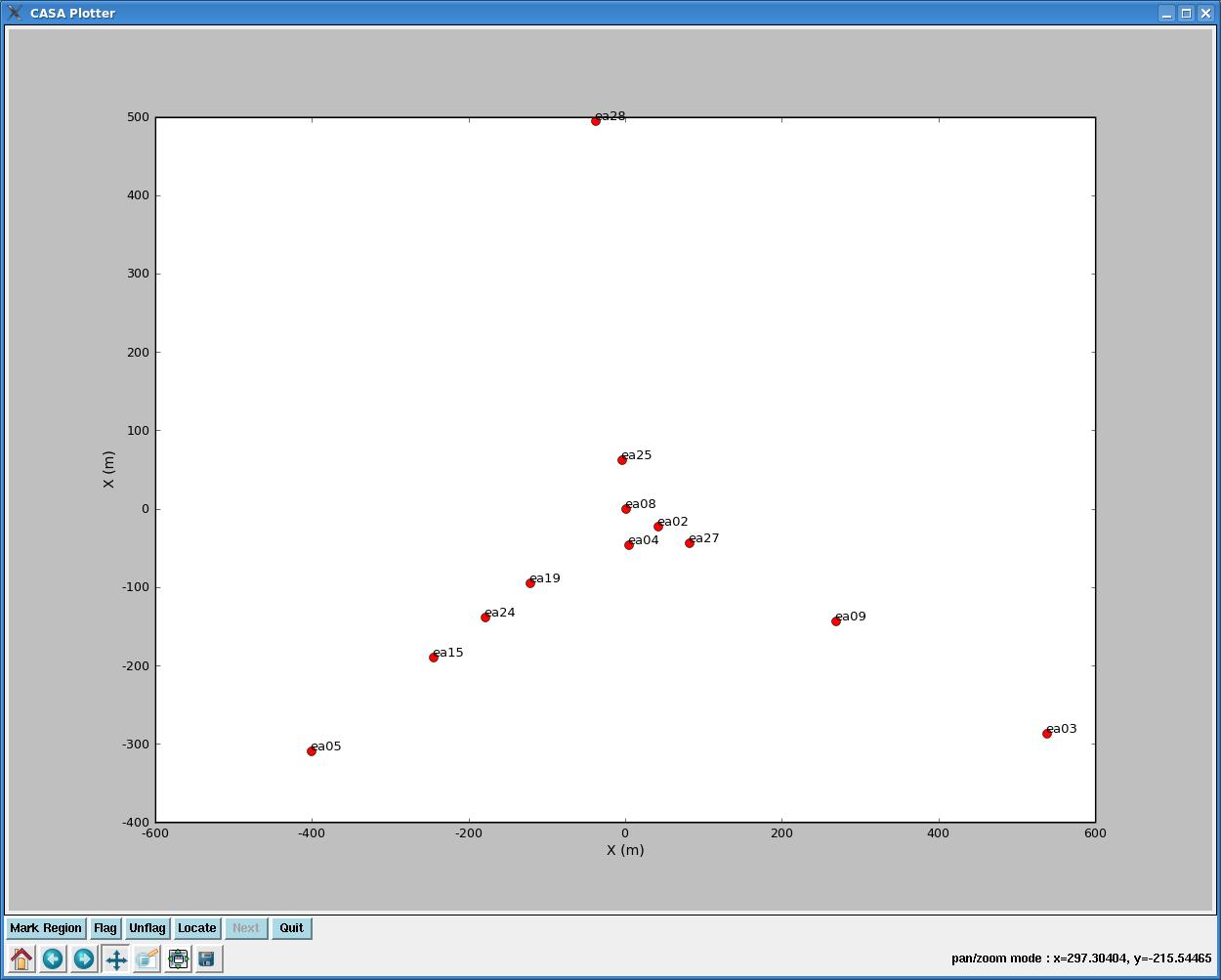
We'd like our reference antenna to be near the center of the array, and the easiest way to envision the array layout is with plotants.
# plotants :: Plot the antenna distribution in the local reference frame:
vis = 'leotest.ms' # Name of input visibility file (MS)
figfile = '' # Save the plotted figure to this file
async = False # If true the taskname must be started using
# plotants(...)
Plotants will produce a plot that looks like this:
You can zoom and pan if you click on this icon: ![]() . Then, holding down your left mouse button and fiddling with the mouse will let you pan; holding down the right mouse button and fiddling with the mouse enables zooming in and out. Make note of an antenna near the center of the array, and check the observing logs to make sure nothing fishy happened to it during the observations. You can also take a quick look at the antenna in plotms to make sure it looks reasonably steady (check out this tutorial for an introduction to plotms).
. Then, holding down your left mouse button and fiddling with the mouse will let you pan; holding down the right mouse button and fiddling with the mouse enables zooming in and out. Make note of an antenna near the center of the array, and check the observing logs to make sure nothing fishy happened to it during the observations. You can also take a quick look at the antenna in plotms to make sure it looks reasonably steady (check out this tutorial for an introduction to plotms).
Here, we choose antenna ea25. When giving this antenna as an input to calibration routines, it can be referred to in several ways: 'ea25', '9', or 'N02' (see the output from listobs).
First Round of Data Flagging
Presently, no flags whatsoever are automatically applied to EVLA data. To apply online flags (for when the telescope is not on source or when the subreflector has not settled), you'll have to use the flagant.py script; see here for directions.
After applying the online flags, check your calibrator data to see if the first few integrations of each scan are bad. If they are, flag them with flagdata in 'quack' mode. This is what the default flagdata parameters look like in our CASA window:
Tip: You'll notice that some parameters are colored differently than others. Standard parameters (familiar from AIPS) are simple black text (like vis). Parameters which are shaded grey (like selectdata) are expandable, and once they have been expanded (like mode), green sub-parameters (like autocorr) will appear. Note that if you were to change mode from 'manualflag' to, say, 'quack', a different menu of green sub-parameters would appear.
To flag the first 10 seconds of each scan, set the flagdata parameters to:
# flagdata :: All purpose flagging task based on selections
vis = 'leotest.ms' # Name of file to flag
flagbackup = True # Automatically back up the current flags?
mode = 'quack' # Mode (manualflag,shadow,quack,summary,autoflag,rfi)
autocorr = False # Flag autocorrelations
unflag = False # Unflag the data specified
quackinterval = 10 # Quack n seconds from scan beginning/end
quackmode = 'beg' # Quack mode. 'beg' ==> beginning of scan. 'endb' ==> end of
# scan. 'end' ==> all but end of scan. 'tail' ==> all but
# beginning of scan
quackincrement = False # Flag incrementally in time?
spw = '' # spectral-window/frequency/channel
field = '' # Field names or field index numbers: ''==>all,
# field='0~2,3C286'
selectdata = False # More data selection parameters (antenna, timerange etc)
async = False # If true the taskname must be started using flagdata(...)
Currently, antenna shadowing is not included in the online flags, so you might have some shadowed antennas in your data. You can also use flagdata to edit these out, with mode= 'shadow'.
# flagdata :: All purpose flagging task based on selections
vis = 'leotest.ms' # Name of file to flag
flagbackup = True # Automatically back up the current flags?
mode = 'shadow' # Mode (manualflag,shadow,quack,summary,autoflag,rfi)
diameter = 28.0 # Effective diameter (m) to use. -1 ==>antenna diameter
spw = '' # spectral-window/frequency/channel
field = '' # Field names or field index numbers: ''==>all, field='0~2,3C286'
selectdata = False # More data selection parameters (antenna, timerange etc)
async = False # If true the taskname must be started using flagdata(...)
We conservatively use a diameter which is a bit bigger than the actual 25m diameter of the EVLA antennae.
Finally, you might want to use flagdata to flag antennas with no receivers or other serious issues. Here is an example where we flag antenna ea13.
# flagdata :: All purpose flagging task based on selections
vis = 'leotest.ms' # Name of file to flag
flagbackup = True # Automatically back up the current flags?
mode = 'manualflag' # Mode (manualflag,shadow,quack,summary,autoflag,rfi)
autocorr = False # Flag autocorrelations
unflag = False # Unflag the data specified
clipexpr = '' # Expression to clip on
clipminmax = [] # Range to use for clipping
clipcolumn = 'DATA' # Data column to use for clipping
clipoutside = True # Clip outside the range, or within it
channelavg = False # Average over channels
spw = '' # spectral-window/frequency/channel
field = '' # Field names or field index numbers: ''==>all, field='0~2,3C286'
selectdata = True # More data selection parameters (antenna, timerange etc)
antenna = 'ea13' # antenna/baselines: ''==>all, antenna = '3,VA04'
timerange = '' # time range: ''==>all, timerange='09:14:0~09:54:0'
correlation = '' # Select data based on correlation
scan = '' # scan numbers: ''==>all
feed = '' # multi-feed numbers: Not yet implemented
array = '' # (sub)array numbers: ''==>all
uvrange = '' # uv range: ''==>all; uvrange = '0~100klambda', default
# units=meters
async = False # If true the taskname must be started using flagdata(...)
Now is also the time to flag any obviously aberrant data. You might choose to flag with viewer, which displays a raster image of your uv data and is similar to SPFLG and TVFLG in AIPS (click here for an introduction to viewer). Alternatively, you can flag using a 2-dimensional interactive plot with plotms (click here for an introduction to plotms). The plotting capabilities can be likened to UVPLT in AIPS, and data points can be flagged (like WIPER in AIPS).
Calibration
Set the Flux Scale
Use the task setjy to find the flux density of the flux calibrator (as in AIPS). In this case, the flux calibrator is 1331+305.
# setjy :: Fills the model column with the visibility of a calibrator vis = 'leotest.ms' # Name of input visibility file field = '1331+305' # One Field name spw = '' # Spectral window identifier (list) modimage = '/home/casa/data/nrao/VLA/CalModels/3C286_L.im' # File location for field model fluxdensity = -1 # Specified flux density [I,Q,U,V]; -1 will lookup values standard = 'Perley-Taylor 99' # Flux density standard async = False # If true the taskname must be started using setjy(...)
In most cases, you should be wary that the flux calibrator could be resolved, and you should use a model (set by the modimage parameter above). CASA comes pre-packed with model images, but finding them might be a bit tricky. Here are some probable paths (see the CASA Cookbook for more detail):
- Red Hat Linux RPMs (RHE4, Fedora 6): /usr/lib/casapy/data/nrao/VLA/CalModels
- MAC OSX .dmg: /opt/casa/data/nrao/VLA/CalModels
- NRAO-AOC stable: /home/casa/data/nrao/VLA/CalModels
Setjy will copy this model to the 'model' data column of your measurement set, and use it in subsequent calibrations of '1331+305'.
You should also note that setjy expects your flux calibrator to be named in certain conventions (See Table 4.1 of the CASA Cookbook for acceptable names). If your observations don't follow these conventions, you'll probably have to change the flux calibrator's field name.
Setjy should output something that looks like this:
2010-02-11 23:57:53 INFO setjy ########################################## 2010-02-11 23:57:53 INFO setjy ##### Begin Task: setjy ##### 2010-02-11 23:57:53 INFO setjy::::casa 2010-02-11 23:57:53 INFO setjy 1331+305 spwid= 0 [I=14.75, Q=0, U=0, V=0] Jy, (Perley-Taylor 99) 2010-02-11 23:57:54 INFO setjy Using model image /home/casa/data/nrao/VLA/CalModels/3C286_L.im 2010-02-11 23:57:54 INFO setjy The model image's reference pixel is 0.000254726 arcsec from 1331+305's phase center. 2010-02-11 23:57:54 INFO setjy Scaling model image to I=14.7461 Jy for visibility prediction. 2010-02-11 23:57:54 INFO setjy Selecting data 2010-02-11 23:57:54 INFO setjy Selected 4818 out of 70290 visibilities. 2010-02-11 23:57:54 INFO setjy Fourier transforming: replacing MODEL_DATA column 2010-02-11 23:57:54 INFO setjy Performing interferometric gridding with convolution function SF 2010-02-11 23:57:55 INFO setjy::::casa 2010-02-11 23:57:55 INFO setjy ##### End Task: setjy ##### 2010-02-11 23:57:55 INFO setjy ##########################################
Note that it has determined a flux of 14.76 Jy for 1331+305. One of the most common signals that something has gone wrong is if setjy claims I=1 Jy.
Calibrate the Bandpasses
Let's use the flux calibrator to calibrate the bandpasses with the task bandpass (similar to BPASS in AIPS). Note that currently, some EVLA antennae may show phase wraps across the bandpass due to delays. If you were using AIPS, you might use FRING to calculate and correct for these delays, but no equivalent task exists in this release of CASA. However, the CASA bandpass calibration should be able to handle phase wraps, so no explicit delay correction is really necessary.
To optimize our bandpass calibration, we'll first need to do a preliminary phase calibration on 1331+305 with gaincal (analogous to CALIB in AIPS).
# gaincal :: Determine temporal gains from calibrator observations
vis = 'leotest.ms' # Nome of input visibility file
caltable = 'leotest_0.gcal' # Name of output gain calibration table
field = '1331+305' # Select field using field id(s) or field name(s)
spw = '*:118~137' # Select spectral window/channels
selectdata = False # Other data selection parameters
solint = 'inf' # Solution interval: egs. 'inf', '60s' (see help)
combine = '' # Data axes which to combine for solve (scan, spw, and/or
# field)
preavg = -1.0 # Pre-averaging interval (sec) (rarely needed)
refant = 'ea25' # Reference antenna name. ' '= '0'
minblperant = 4 # Minimum baselines _per antenna_ required for solve
minsnr = 0.0 # Reject solutions below this SNR
solnorm = False # Normalize average solution amplitudes to 1.0 (G, T only)
gaintype = 'G' # Type of gain solution (G, T, or GSPLINE)
calmode = 'p' # Type of solution" ('ap', 'p', 'a')
append = False # Append solutions to the (existing) table
gaintable = [''] # Gain calibration table(s) to apply on the fly
gainfield = [''] # Select a subset of calibrators from gaintable(s)
interp = [''] # Temporal interpolation for each gaintable (=linear)
spwmap = [] # Spectral windows combinations to form for gaintables(s)
gaincurve = False # Apply internal VLA antenna gain curve correction
opacity = 0.0 # Opacity correction to apply on the fly (nepers)
parang = False # Apply parallactic angle correction on the fly
async = False # If true the taskname must be started using gaincal(...)
You'll want to find an intuitive naming convention for your calibration tables; here, we output the gain calibration to a table called 'leotest_0.gcal'. The above parameters calibrate only the phases, not amplitudes (calmode= 'p') for the flux calibrator (field= '1331+305')'. We use a narrow channel range in the center of the bandpass (spw= '*:118~137'; see this description for more detail on selecting spectral windows and channels), chosen so that the bandpass does not vary much across this range. The combination of parameters solint= 'inf' and combine= ' ' tell gaincal to time-integrate across an entire calibrator scan in finding a solution. We have assigned a source model to the flux calibrator with setjy, so we do not need to set a uvrange. Don't forget to set the reference antenna (refant= 'ea25').
The calibration tables will be outputted as a directory in your present working directory. If you type 'ls' on the CASA command line, you should see a directory named for your caltable.
Tip: If, for some reason, you'd like to delete this caltable, the neatest way to do it is: rmtables('leotest_1.bcal') If you get an error when running rmtables, it is likely because CASA is still accessing the table. If you really want to delete the table, you'll have to either (a) quit out of CASA and then restart it or (b) try the clumsier rm -rf leotest_1.bcal.
Check that the phase calibration solutions look reasonable with plotcal (which, in this case, is similar to SNPLT in AIPS).
# plotcal :: An all-purpose plotter for calibration results caltable = 'leotest_0.gcal' # Name of input calibration table xaxis = '' # Value to plot along x axis (time,chan,freq...see pdoc) yaxis = 'phase' # Value to plot along y axis (amp,phase,real,imag,snr,antenna) poln = '' # Antenna polarization to plot (RL,R,L,XY,X,Y,/) field = '' # field names or index of calibrators: ''==>all antenna = '' # antenna/baselines: ''==>all, antenna = '3,VA04' spw = '' # spectral window:channels: ''==>all, spw='1:5~57' timerange = '' # time range: ''==>all subplot = 111 # Panel number on display screen (yxn) overplot = False # Overplot solutions on existing display clearpanel = 'Auto' # Specify if old plots are cleared or not (ignore) iteration = '' # Iterate plots on antenna,time,spw,field plotrange = [] # plot axes ranges: [xmin,xmax,ymin,ymax] showflags = False # If true, show flagged solutions plotsymbol = 'o' # pylab plot symbol plotcolor = 'blue' # initial plotting color markersize = 5.0 # Size of plotted marks fontsize = 10.0 # Font size for labels showgui = True # Show plot on gui figfile = '' # ''= no plot hardcopy, otherwise supply name async = False # If true the taskname must be started using plotcal(...)
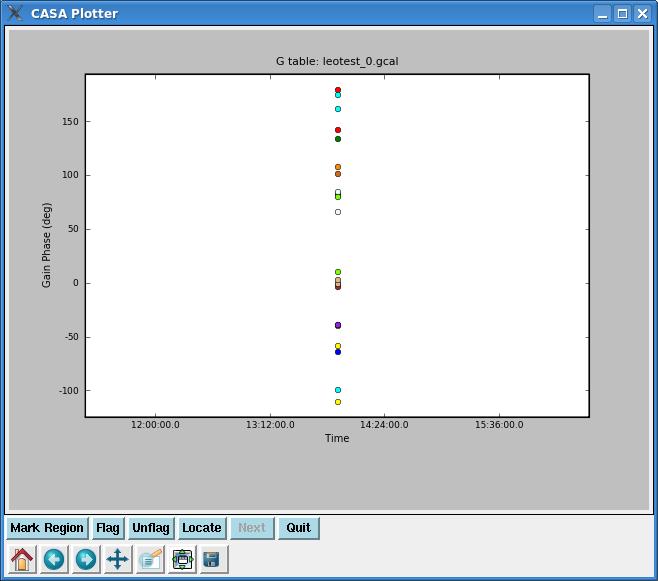
Plotcal with the above parameters will plot phase vs. time for all antennae overplotted on one another. However, in this specific example, because there is only one flux calibrator scan and because we are time-integrating across it, there should only be one point per antenna (see figure at right).
Now, we're ready to solve for bandpass solutions with bandpass:
# bandpass :: Calculates a bandpass calibration solution
vis = 'leotest.ms' # Nome of input visibility file
caltable = 'leotest_1.bcal' # Name of output gain calibration table
field = '1331+305' # Select field using field id(s) or field name(s)
spw = '' # Select spectral window/channels
selectdata = False # Other data selection parameters
solint = 'inf' # Solution interval
combine = 'scan' # Data axes which to combine for solve (scan, spw, and/or field)
refant = 'ea25' # Reference antenna name
minblperant = 4 # Minimum baselines _per antenna_ required for solve
solnorm = True # Normalize average solution amplitudes to 1.0 (G, T only)
bandtype = 'B' # Type of bandpass solution (B or BPOLY)
fillgaps = 0 # Fill flagged solution channels by interpolation
append = False # Append solutions to the (existing) table
gaintable = 'leotest_0.gcal' # Gain calibration table(s) to apply on the fly
gainfield = [''] # Select a subset of calibrators from gaintable(s)
interp = [''] # Interpolation mode (in time) to use for each gaintable
spwmap = [] # Spectral windows combinations to form for gaintables(s)
gaincurve = False # Apply internal VLA antenna gain curve correction
opacity = 0.0 # Opacity correction to apply (nepers)
parang = False # Apply parallactic angle correction
async = False # If true the taskname must be started using bandpass(...)
Important parameters to consider are:
- caltable: the name of the bandpass calibration table that will be generated
- field: the name of your bandpass calibrator
- refant: the reference antenna name
- solnorm: =True. Important to set! We want the bandpass solutions to be normalized because we don't want bandpass trying to calibrate the amplitudes.
- gaintable: the name of your preliminary phase calibration table, which bandpass will apply on the fly.
- solint and combine: the bandpass solution time interval. The above settings will average all data on the bandpass calibrator together in time, even across scan boundaries (combine= 'scan').
- bandtype: Because your shared-risk EVLA data likely has some significant delays in it, we strongly suggest that you use bandtype= 'B' (as a polynomial solution 'BPOLY' will not be able to handle phase wraps across your bandpass).
After running bandpass and getting some helpful output in the CASA logger, we can also plot up our bandpass solutions with plotcal (which, in this case, can be likened to POSSM in AIPS with APARM(8)=2). Plotcal parameters that look like this:
# plotcal :: An all-purpose plotter for calibration results caltable = 'leotest_1.bcal' # Name of input calibration table xaxis = '' # Value to plot along x axis (time,chan,freq...see pdoc) yaxis = '' # Value to plot along y axis (amp,phase,real,imag,snr,antenna) poln = '' # Antenna polarization to plot (RL,R,L,XY,X,Y,/) field = '' # field names or index of calibrators: ''==>all antenna = '' # antenna/baselines: ''==>all, antenna = '3,VA04' spw = '' # spectral window:channels: ''==>all, spw='1:5~57' timerange = '' # time range: ''==>all subplot = 111 # Panel number on display screen (yxn) overplot = False # Overplot solutions on existing display clearpanel = 'Auto' # Specify if old plots are cleared or not (ignore) iteration = 'antenna' # Iterate plots on antenna,time,spw,field plotrange = [] # plot axes ranges: [xmin,xmax,ymin,ymax] showflags = False # If true, show flagged solutions plotsymbol = 'o' # pylab plot symbol plotcolor = 'blue' # initial plotting color markersize = 5.0 # Size of plotted marks fontsize = 10.0 # Font size for labels showgui = True # Show plot on gui figfile = '' # ''= no plot hardcopy, otherwise supply name async = False # If true the taskname must be started using plotcal(...)
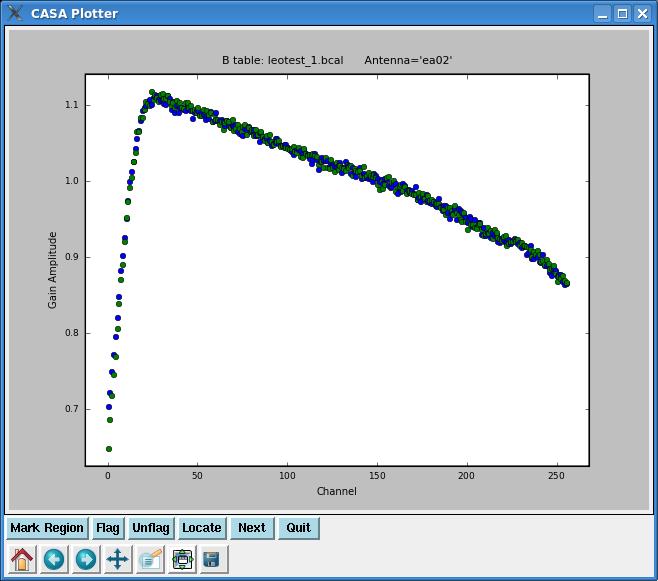
will plot the bandpass solutions (amplitude vs. channel) for each antenna individually. Note that if we had left iteration as its default:
iteration = '' # Iterate plots on antenna,time,spw,field
bandpasses for all antennae would be overplotted in the same window. By setting iteration= 'antenna', plotcal will plot bandpasses for each antenna separately. The plot can be advanced to the next antenna by clicking the ![]() button.
button.
If you want to plot the bandpass phase solutions, simply set yaxis= 'phase'. If you only want to plot one polarization, set the poln parameter. It is also possible to place multiple plots in one window using the subplot parameter; see the CASA Cookbook for more details.
Calibrate the Gains
Now it's time for the amplitude and phase calibration with gaincal. If you are going to use the same uvrange for all calibrator sources, then you can run gaincal simultaneously on them all. Check with the calibrator manual to see the acceptable uvrange for each of your calibrators (if you have assigned a source model to your flux calibrator with setjy, then for that source you can ignore the suggested uvrange in the calibrator manual and use the full range). Here our phase calibrator (J1042+1203) has a suggested uvrange of 0--5 kilolambda, so we are going to run gaincal separately for the flux calibrator and the phase calibrator.
# gaincal :: Determine temporal gains from calibrator observations
vis = 'leotest.ms' # Nome of input visibility file
caltable = 'leotest_1.gcal' # Name of output gain calibration table
field = '1331+305' # Select field using field id(s) or field name(s)
spw = '*:25~250' # Select spectral window/channels
selectdata = False # Other data selection parameters
solint = 'inf' # Solution interval: egs. 'inf', '60s' (see help)
combine = '' # Data axes which to combine for solve (scan, spw, and/or field)
preavg = -1.0 # Pre-averaging interval (sec) (rarely needed)
refant = 'ea25' # Reference antenna name. ' '= '0'
minblperant = 4 # Minimum baselines _per antenna_ required for solve
minsnr = 0.0 # Reject solutions below this SNR
solnorm = False # Normalize average solution amplitudes to 1.0 (G, T only)
gaintype = 'G' # Type of gain solution (G, T, or GSPLINE)
calmode = 'ap' # Type of solution" ('ap', 'p', 'a')
append = False # Append solutions to the (existing) table
gaintable = 'leotest_1.bcal' # Gain calibration table(s) to apply on the fly
gainfield = [''] # Select a subset of calibrators from gaintable(s)
interp = [''] # Temporal interpolation for each gaintable (=linear)
spwmap = [] # Spectral windows combinations to form for gaintables(s)
gaincurve = False # Apply internal VLA antenna gain curve correction
opacity = 0.0 # Opacity correction to apply on the fly (nepers)
parang = False # Apply parallactic angle correction on the fly
async = False # If true the taskname must be started using gaincal(...)
The above parameters calibrate the amplitudes and phases (calmode= 'ap') for the flux calibrator (field= '1331+305') and outputs the gain calibration to a table called 'leotest_1.gcal'. We only use channels 25--250 to calibrate (spw= '*:25~250'), chosen by visual inspection of the bandpass solutions with plotcal. Gaincal will apply the bandpass calibration ('leotest_1.bcal') to these channels, average them together, and then find gain solutions. We use one solution time interval per scan (solint= 'inf' and combine= ' '). In order for flux bootstrapping to work properly in the next calibration step, we want to refrain from normalizing the solutions (solnorm= False). Don't forget to set the reference antenna (refant= 'ea25') and apply the bandpass calibration (gaintable= 'leotest_1.bcal').
Next, we will calibrate the phase calibrator (field= 'J1042+1203') and set a limited uvrange (uvrange= '0~5klambda'):
# gaincal :: Determine temporal gains from calibrator observations
vis = 'leotest.ms' # Nome of input visibility file
caltable = 'leotest_1.gcal' # Name of output gain calibration table
field = 'J1042+1203' # Select field using field id(s) or field name(s)
spw = '*:25~250' # Select spectral window/channels
selectdata = True # Other data selection parameters
timerange = '' # Select data based on time range
uvrange = '0~5klambda' # Select data within uvrange (default units meters)
antenna = '' # Select data based on antenna/baseline
scan = '' # Scan number range
msselect = '' # Optional complex data selection (ignore for now)
solint = 'inf' # Solution interval: egs. 'inf', '60s' (see help)
combine = '' # Data axes which to combine for solve (scan, spw, and/or field)
preavg = -1.0 # Pre-averaging interval (sec) (rarely needed)
refant = 'ea25' # Reference antenna name. ' '= '0'
minblperant = 4 # Minimum baselines _per antenna_ required for solve
minsnr = 0.0 # Reject solutions below this SNR
solnorm = False # Normalize average solution amplitudes to 1.0 (G, T only)
gaintype = 'G' # Type of gain solution (G, T, or GSPLINE)
calmode = 'ap' # Type of solution" ('ap', 'p', 'a')
append = True # Append solutions to the (existing) table
gaintable = 'leotest_1.bcal' # Gain calibration table(s) to apply on the fly
gainfield = [''] # Select a subset of calibrators from gaintable(s)
interp = ['linear'] # Temporal interpolation for each gaintable (=linear)
spwmap = [] # Spectral windows combinations to form for gaintables(s)
gaincurve = False # Apply internal VLA antenna gain curve correction
opacity = 0.0 # Opacity correction to apply on the fly (nepers)
parang = False # Apply parallactic angle correction on the fly
async = False # If true the taskname must be started using gaincal(...)
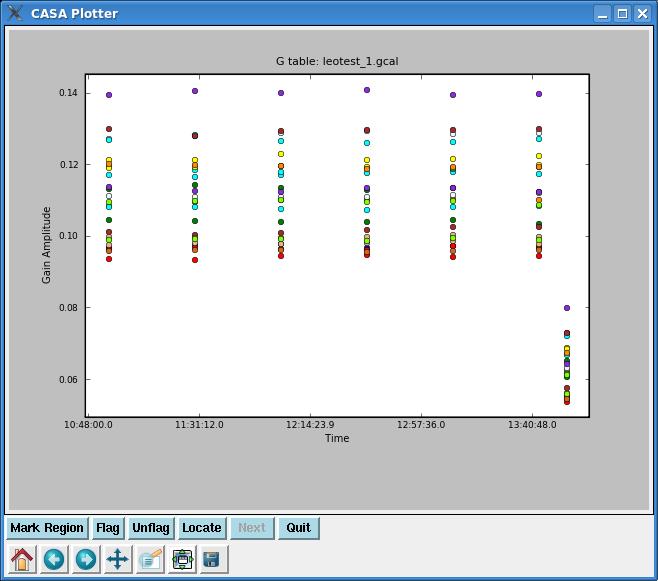
Note that we appended this calibration (append= True) to the original calibration table (caltable= 'leotest_1.gcal').
We'll check our gain solutions with plotcal. If we give it a gain table, the default plot will be amplitude vs. time. The plot for our calibration can be seen to the right, with all antennae overplotted in different colors. You'll also want to check phase vs. time.
Apply the Flux Scale
Next, we'll bootstrap the flux scale that was found by running setjy onto our phase calibrator with the CASA task fluxscale (synonymous with GETJY in AIPS).
# fluxscale :: Bootstrap the flux density scale from standard calibrators vis = 'leotest.ms' # Name of input visibility file (MS) caltable = 'leotest_1.gcal' # Name of input calibration table fluxtable = 'leotest_1.fluxscale' # Name of output, flux-scaled calibration table reference = '1331+305' # Reference field name(s) (transfer flux scale FROM) transfer = 'J1042+1203' # Transfer field name(s) (transfer flux scale TO), '' -> all append = True # Append solutions? refspwmap = [-1] # Scale across spectral window boundaries. See help fluxscale async = False # If true the taskname must be started using fluxscale(...)
We get output like this:
2010-02-14 21:35:04 INFO fluxscale ########################################## 2010-02-14 21:35:04 INFO fluxscale ##### Begin Task: fluxscale ##### 2010-02-14 21:35:04 INFO fluxscale::::casa 2010-02-14 21:35:04 INFO fluxscale Opening MS: leotest.ms for calibration. 2010-02-14 21:35:04 INFO fluxscale Initializing nominal selection to the whole MS. 2010-02-14 21:35:04 INFO fluxscale Beginning fluxscale--(MSSelection version)------- 2010-02-14 21:35:04 INFO fluxscale Found reference field(s): 1331+305 2010-02-14 21:35:04 INFO fluxscale Found transfer field(s): J1042+1203 2010-02-14 21:35:04 INFO fluxscale Flux density for J1042+1203 in SpW=0 is: 3.09102 +/- 0.0129722 (SNR = 238.28, nAnt= 12) 2010-02-14 21:35:04 INFO fluxscale Appending result to leotest_1.fluxscale 2010-02-14 21:35:04 INFO fluxscale Appending solutions to table: leotest_1.fluxscale 2010-02-14 21:35:04 INFO fluxscale::::casa 2010-02-14 21:35:04 INFO fluxscale ##### End Task: fluxscale #####
which tells us that our phase calibrator has a flux density of 3.1 Jy. Here, it is good to be on guard for flux densities which are suspiciously close to 1 Jy or errors in the derived flux density which are very large. Note that, unlike in AIPS, fluxscale writes a new calibration table, and this is what we will want to apply to our data. The output table 'leotest_1.fluxscale' does not only include information about the flux density scale, but also includes the full solution of gains, copied over from 'leotest_1.gcal'---so it supersedes 'leotest_1.gcal'.
Apply the Calibration
Now, we're happy with our calibration and ready to apply it to the data with applycal. You'll want to give applycal all relevant calibration tables! Here we are going to calibrate bandpasses, gains, and flux densities.
Applycal might be compared with CLCAL in AIPS, except that instead of writing a new calibration (CL) table, applycal will write a new calibrated column of data in your '.ms' file. It also differs from CLCAL in that we will use applycal to apply all calibrations, not only gain/flux density solutions. After we run applycal, we will still be able to access the raw data (called 'data' by many tasks), but there will also be calibrated data (called 'corrected' by many tasks).
# applycal :: Apply calibrations solutions(s) to data vis = 'leotest.ms' # Nome of input visibility file field = '' # Select field using field id(s) or field name(s) spw = '' # Select spectral window/channels selectdata = False # Other data selection parameters gaintable = ['leotest_1.bcal', 'leotest_1.fluxscale'] # Gain calibration table(s) to apply on the fly gainfield = [''] # Select a subset of calibrators from gaintable(s) interp = 'linear' # Temporal Interpolation type. default=linear spwmap = [] # Spectral windows combinations to form for gaintables(s) gaincurve = False # Apply internal VLA antenna gain curve correction opacity = 0.0 # Opacity correction to apply (nepers) parang = False # Apply parallactic angle correction calwt = True # Calibrate data weights from all relevant calibrations async = False # If true the taskname must be started using applycal(...)
The above settings for applycal will calibrate all sources, linearly interpolating between gaincal solutions. It will apply the flux, gain, and bandpass calibration (gaintable= ['leotest_1.bcal', 'leotest_1.fluxscale']).
Inspect the Calibrated Data
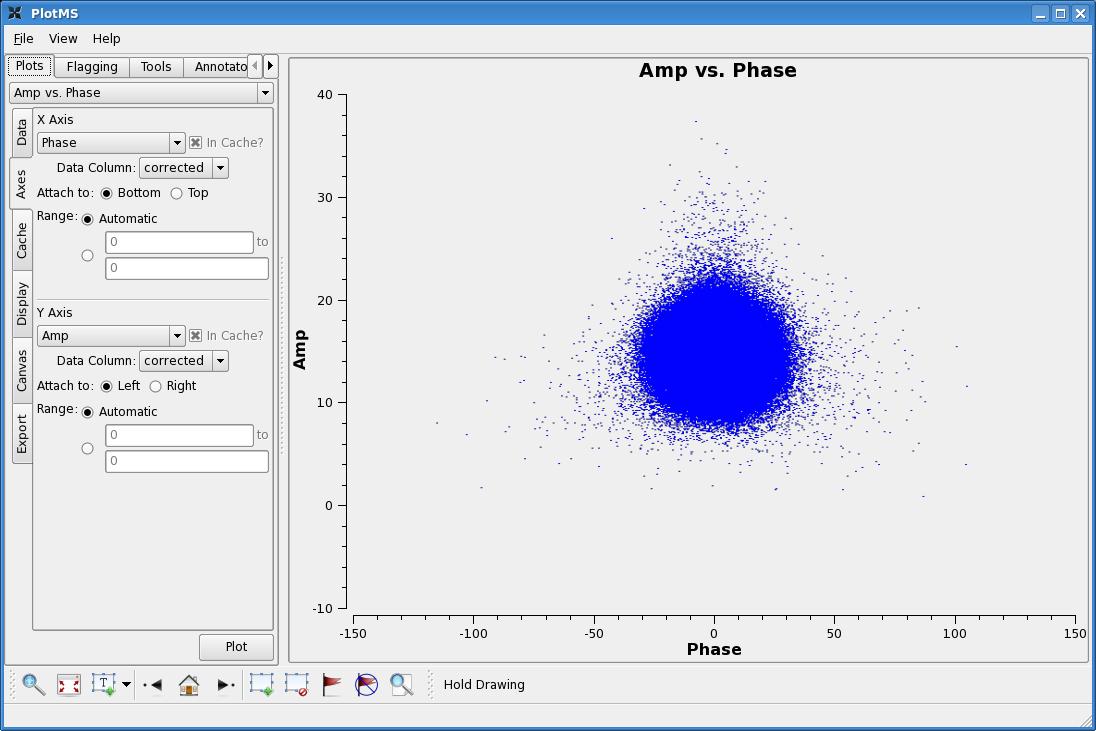
A good way to check our calibration is to plot up amplitudes and phases for our calibrated calibrators. By default plotms plots up the raw data column of a '.ms' file. If, at this point, we were to leave the plotms parameters as their defaults and plot up amplitude vs. phase for 1331+305, we'd see a plot that looks like this:
To plot the calibrated data, go to the plotms window and click on the Axes tab. You will see drop-down menus called Data Column. Select 'corrected', and the plot should now look like this:
A nice ball centered in Amplitude around the expected flux density (found by setjy and centered in Phase around zero degrees, just as we'd expect for a calibrated point source.
You can also easily switch between raw and calibrated data in viewer by selecting a Visibility Type, which can be found in MS and Visibility Selection section of the Data Display Options window. There will be a choice of 'observed' or 'corrected' visibilities. You can even display the 'model' and 'residual' visibilities.
After calibration, new bad data will sometimes pop up. If there are a lot of bad data or the bad points are severely aberrant, you might want to take this opportunity to flag them and then recalibrate the data. In this case, it is probably good form to delete the corrected data column with clearcal (but note that this task can be quite slow, and this step may be unnecessary as gaincal and bandpass will, regardless, use the raw data, and applycal will overwrite the old 'corrected' data if it is run again). Then start again from the bandpass step above (making sure to give calibration tables different names).
Split Off the Science Targets
Finally, we'll want to split off our calibrated science targets from the multi-source measurement set, so that we can image them! Use split, as you would in AIPS (However, note that split in CASA can not apply calibration as AIPS SPLIT does. You have to first apply the calibration with applycal, the specify datacolumn= 'corrected').
# split :: Create a visibility subset from an existing visibility set vis = 'leotest.ms' # Name of input measurement set outputvis = 'leo1_split.ms' # Name of output measurement set datacolumn = 'corrected' # Which data column(s) to split out field = 'Leo-1' # Select field using field id(s) or field name(s) spw = '' # Select spectral window/channels width = '1' # Number of channels to average to form one output channel antenna = '' # Select data based on antenna/baseline timebin = '0' # Value for timeaveraging timerange = '' # Select data based on time range scan = '' # select data based on scan numbers array = '' # Select (sub)array by array ID number(s) uvrange = '' # select data based on uv distance range async = False # If true the taskname must be started using split(...)
The above will split off the calibrated data (datacolumn= 'corrected') for the first Leo field (field= 'Leo-1'). We'll want to repeat the above for the second Leo pointing, 'Leo-2'.
Now is also the time to note that all EVLA OSRO datasets are currently observed with 1 second integrations. This is likely to be more time resolution than you need for imaging, and your data set is probably quite large, making it time-consuming to reduce. Therefore, you might consider time averaging by increasing the timebin parameter from '0s' to your desired value). See the EVLA Observational Status Summary for advice on time averaging.
Now we are ready to flag these science targets and image them! Continue on to this imaging tutorial for the next installment....
Last checked on CASA Version 3.0.0 (r9860).
--Laura Chomiuk 23:35, 14 February 2010 (UTC)