ATCA CABB Continuum Tutorial
(This Page is Currently Under Development)
About This Tutorial
This tutorial provides an example of how to process a continuum observation from the Australian Telescope Compact Array (ATCA) using CASA. Special consideration is given to the calibration and imaging requirements of broadband observations with the Compact Array Broadband Backend (CABB). Note that this tutorial has been adapted from the MIRIAD tutorial written for the same data set by Jamie Stephens: http://www.atnf.csiro.au/computing/software/miriad/tutorials.html.
The Australian Telescope Compact Array
The ATCA is an array of six 22 m diameter antennas located 237 m above sea level at latitude -30° 18′ 46.385″ south, longitude 149° 33′ 00.500″ east. The array has a 3 km east-west track with a 214 m northern spur. Five antennas can be moved along these tracks, with the sixth antenna at a fixed position 3 km to the west of the east-west track. The longest possible baseline is, therefore, 6 km. The array can be used for observations in five wavelength bands between 27 cm and (with five antennas only) 3 mm, between frequencies of approximately 1.1 GHz and 105 GHz. More information about ATCA is available here: http://www.narrabri.atnf.csiro.au
About These Observations
The data used here is from the ATCA project CX208. This project's goal was to measure continuum properties of a source that appeared to have a very steep spectral index at low frequencies (below 1 GHz). The ATCA 16 cm system (which covers the 1.1 - 3.1 GHz range) was used to observe this source on April 28, 2011. The array was in the 6A configuration at the time, because it was thought that confusion may have been a possible explanation for the apparent spectral steepness.
Obtaining The Data
The data can be downloaded from the Australian Telescope Online Archive (ATOA) here: http://atoa.atnf.csiro.au. Search for project code cx208 and select the search result titled 2011-04-28_1858.CX208.
Importing The Data
At present, CASA does not have a task to import the RPFITS format used by the ATCA. As such, data from the ATCA archive must first be loaded with MIRIAD (http://www.atnf.csiro.au/computing/software/miriad/) using the MIRIAD task atlod:
inp atlod
in = 2011-04-28_1858.CX208
out = cx208.uv
ifsel = 1
options = birdie,rfiflag,noauto,xycorr
These data can then be converted to measurement set format using the CASA task importmiriad. Note that this step requires CASA version 4.3 or later.
inp importmiriad
mirfile = 'cx208.uv'
vis = 'cx208.ms'
Summarizing the Observation
The task listobs will summarize key aspects of the measurement set. Enter the following lines in the CASA terminal window:
inp listobs
vis=’cx208.ms’
The summary will be written to the log file, which we can view with the Logger window. We see that this observation consists of data from 6 antennas, which have observed 3 fields for a total of 29 scans, and that the data has been recorded in one spectral window with 2049 channels. Note that each of these elements can be referenced using a zero-based index. Scan 0 refers to the 1934-638 field (field ID 0) which we will use for bandpass and flux calibration. Field ID 1 is the 2058-425 field, which we will use for complex gain calibration. From the scan listing, we see that short scans of field 1 bracket longer scans of field 2051-377 (field ID 2), which is our science target.
Inspecting the Visibilities
We will use the task plotms to plot the UV data for the flux calibrator field:
inp plotms
field=’0’
Note that the ‘vis’ parameter has been carried over from the previous task automatically. By default, plotms will show the amplitude versus time. Lets switch to a plot of amplitude vs. frequency. Select the ‘Axes’ tab on the left of the plotms window, then under ‘X Axis’ use the dropdown menu to select ‘Frequency.’ Click ‘Plot’ (near the bottom of the plotms window) to refresh the display. The most striking feature of this plot is the large spikes of RFI. Fortunately they are confined to just a few channels and we will flag them in a minute. Another feature of this plot is the set of low amplitude points. These are from the cross-hand visibility products, i.e., XY and YX. Confirm this by coloring the plot points by correlation. Select the ‘Display’ tab, click on the ‘Colorize by’ checkbox, and use the adjacent dropdown menu to select ‘Corr,’ then click ‘Plot.’
Flagging the Flux Calibrator
Although we could flag data interactively using plotms, it is also a good idea to be familiar with CASA’s automatic RFI flagging utilities. We will use the RFLAG algorithm, which clips data with a high vector RMS using sliding windows in time and frequency. Using the cross-hand visibilities can improve the performance of this algorithm, especially when run on uncalibrated data.
inp flagdata
mode=’rflag’
correlation=’xy,yx’
Note that both the ‘vis’ and ‘field’ parameters have been carried over from the previous task. This behavior can be convenient, but its always good to check the inputs carefully before running each task. Return to the plotms window, click on ‘force reload’ and click ‘Plot.’ Most of the interference should now be gone. We can flag the remaining few points interactively in plotms. Click on the ‘Mark Regions’ icon at the bottom of the plotms window (rectangle with green plus sign), draw boxes around the points you want to flag, then click the ‘Flag’ icon (red flag).
Examples Below
The output measurement set, i.e., the data to be worked on, is ag733.ms. Note that the (interesting) output of listobs and vishead go to the CASA message log window.
listobs is similar to LISTR in AIPS and provides a list of scans, sources and their respective indices, and antennas. You can also get a more useful plot of antennas, akin to the output of the AIPS task PRTAN, using plotants. The antennas plot aids the selection of a reference antenna; the antenna VA05 looks like a reasonable choice.
# In CASA
vis = 'ag733.ms'
plotants()
Editing the Measurement Set
This technique is detailed in the tutorial Data flagging with plotms. Here's a shorter version.
# In CASA
plotms()
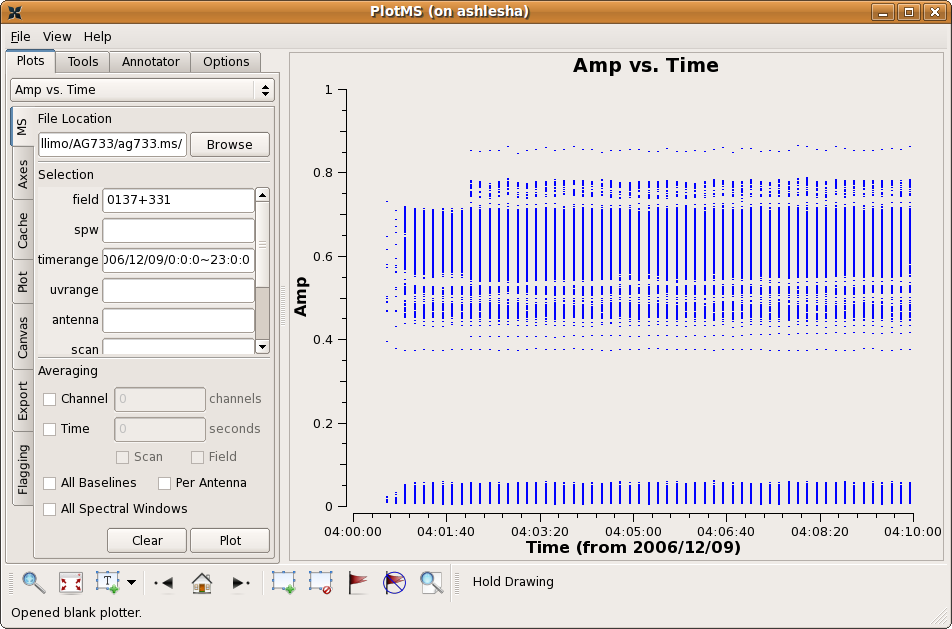
Plotms brings up the following graphical display.
In this example, the measurement set was selected by navigating to and highlighting the measurement set ag733.ms (which will look like a directory to your file browser, because measurement sets are actually directory structures). A specific subset of data (the flux calibrator 3C48 = 0137+331 on the first observing day) was selected by specifying in the appropriate "Data -> Selection" boxes,
- field = 0137+331, and
- timerange = 2006/12/09/0:0:0~23:0:0.
For this specific field, we see no obvious bad data. Note that the low points are actually cross-polarization data; if you want to only look at the right and left polarizations, enter
- corr = RR, LL.
For the sake of this example, we'll clip out the early part of the observations:
Firstly, box the region to be flagged by using the "Mark Regions" button ![]() . Next, go the the "Flag" tab at top, and select "Extend flags" and "Correlation", so that we will delete the RL and LR data as well (despite the fact that we're not plotting it). Then, press the "Go" button, or "Flag" button
. Next, go the the "Flag" tab at top, and select "Extend flags" and "Correlation", so that we will delete the RL and LR data as well (despite the fact that we're not plotting it). Then, press the "Go" button, or "Flag" button ![]() to zap those data.
to zap those data.
The viewer tool provides utilities to flag data in a manner similar to the AIPS task TVFLG; see the tutorial Data flagging with viewer.
If we leave the field and timerange filter boxes blank and plot again, we see a number of visibilities with extremely large amplitudes. By highlighting and locating the bad points (described in detail in the data flagging with plotms tutorial), we find most of the bad data can be attributed to one antenna, 'VA15', at specific times, and one time range where all antennas contribute. In the text below we flag these points using flagcmd and tflagdata; you can also flag data interactively in the plotms display (though this is generally not encouraged since it does not save backup versions of the flags, and it can be easy to flag data by mistake or miss data that's not plotted).
# In CASA
flagcmd(vis='ag733.ms', inpmode='cmd',
command=["antenna='VA15' timerange='2006/12/09/04:45:10.0~04:45:20.0'",
"antenna='VA15' timerange='2006/12/09/06:12:30.0~06:14:10.0'",
"antenna='VA15' timerange='2006/12/13/04:32:40.0~04:35:30.0'",
"antenna='VA15' timerange='2006/12/09/06:12:30.0~06:12:40.0'",
"antenna='VA15' timerange='2006/12/09/06:13:00.0~06:13:40.0'",
"antenna='VA15' timerange='2006/12/09/06:13:50.0~06:14:00.0'",
"antenna='VA15' timerange='2006/12/09/06:14:00.0~06:14:10.0'"],
action='apply')
tflagdata(vis='ag733.ms', timerange='2006/12/09/06:24:50.0~06:25:00.0')
Notice that you can flag in one line multiple time ranges for a given antenna. Plot the data again to make sure the amplitudes look reasonable (if plotms is already open, just hit Shift+Plot to reload with the current flags):
# In CASA
plotms()
Careful with Cross-Pols
By default, plotms plots all of the data, including cross-polarization. All those low-amplitude points in your data are probably not junk. They are real cross-polarization data. Flagging those data will cause grief later on. Take a look at the tutorial data flagging with plotms.
Tread lightly! And, whenever possible, flag with tflagdata and keep careful track of what you flag in a separate file.
A Python Interlude
CASA uses python as an interactive wrapper and scripting language. Python offers the ability to define global variables on the fly, and it's worth using such variables to take care of bookkeeping. First we'll define some list variables that store calibrator and source information.
ampcallist = ['0137+331']
phasecallist = ['2250+143', '0119+321', '0237+288', '0239-025',
'0323+055', '0339-017', '0423-013']
sourcelist = ['NGC7469', 'MRK0993', 'MRK1040', 'NGC1056', 'NGC1068',
'NGC1194', 'NGC1241', 'NGC1320', 'F04385-0828',
'NGC1667']
allcals = ampcallist + phasecallist
print allcals
It will also be helpful later to have a way of referencing which phase calibrator should be assigned to which source. We can define the matching explicitly by using a dictionary variable.
calDict = {'NGC7469':'2250+143',
'MRK0993':'0119+321',
'MRK1040':'0237+288',
'NGC1056':'0237+288',
'NGC1068':'0239-025',
'NGC1194':'0323+055',
'NGC1241':'0323+055',
'NGC1320':'0339-017',
'F04385-0828':'0423-013',
'NGC1667':'0423-013'}
Those variables being defined, we can proceed with the calibration (and not have to type source and calibrator names over and over).
Calibration
Set the Flux Scale
As in AIPS, CASA sets the flux scale using an observation of a flux calibrator (here, 0137+331) and the task setjy. For this example, we'll determine the calibration in reference to a model image of the calibrator source. Look at the inputs of setjy and get it ready to go!
# in CASA
# perhaps restore setjy back to its defaults
default setjy
inp
vis = 'ag733.ms'
field = '0137+331'
# For the model image, need to specify the correct directory, something like the following
#
# load the CASA path from environment variable
import os
casapath = os.environ.get('CASAPATH').split()[0]
# generate full path to the image
modimage = casapath + '/data/nrao/VLA/CalModels/3C48_C.im'
go
That example was set up to demonstrate some AIPS heritage. You could equally well run setjy the following way.
# in CASA
setjy('ag733.ms', field = '0137+331', modimage = casapath + '/data/nrao/VLA/CalModels/3C48_C.im')
Determine calibration solutions
At this stage the data have an overall flux scaling determined, but full gain solutions aren't there yet. The relevant task is gaincal (analogous to the AIPS task CALIB).
Rather than generate solution tables (SN tables in AIPS) that are attached to the measurement set, gaincal will produce a separate table, which we'll call cal.G.
rmtables('cal.G') # get rid of any old, unmeritorious versions of the calibration
#
# Some caution on the following step. The solution interval is set to 'inf,' meaning to average over entire
# scans of each calibrator in turn. If a given scan is however longer than the coherence time (esp. on long baselines),
# some decorrelation will occur and the flux calibration will be thrown off. Suggest, e.g., solint='1min' or solint='2min'
# as reasonable alternatives to solint='inf'. Trial and error may be your friend here.
#
gaincal(vis='ag733.ms',caltable='cal.G', field=','.join(allcals), solint='inf',
refant='VA05', append=False, gaincurve=False)
plotcal('cal.G',yaxis='amp',timerange='2006/12/09/04:0:0~06:30:0')
There's some python trickery buried in this example. If you are unfamiliar with the string method join, try the following example. Recall that we had defined the variable allcals above in A Python Interlude.
print allcals
print ','.join(allcals)
It's worth playing with the timerange, etc. in plotcal to explore the solutions. You may notice that the antenna 'EA26' looks a little junky on the second day. Here's how to flag it manually and then repeat the calibration (be sure to close the plotcal window displaying the previous solutions first).
tflagdata(vis='ag733.ms',antenna='EA26', timerange='2006/12/13/0:0:0~24:0:0')
rmtables('cal.G')
gaincal(vis='ag733.ms',caltable='cal.G', field=','.join(allcals), solint='inf',
refant='VA05', append=False, gaincurve=False)
Go back and re-plot the calibration; this example shows even more AIPS heritage.
tget plotcal
plotcal()
Finally, bootstrap the flux scale of the flux calibrator onto the phase calibrators. AIPS called it GETJY, but CASA calls it fluxscale.
rmtables('cal.Gflx') # remove any old versions of the calibration table, if necessary
myFluxes = fluxscale(vis='ag733.ms', caltable='cal.G', reference=','.join(ampcallist),
transfer=','.join(phasecallist), fluxtable='cal.Gflx', append=False)
Here, the calibration table cal.G is modified and stored as 'cal.Gflx.' The Python dictionary "myFluxes" will also contain information about the flux scaling and other useful tidbits (type "myFluxes" to see its contents). So far, solutions have been generated only for the calibrators, and they have not yet been transfered to the sources.
We could equally well have used reference='0137+337', but use of the join method allows for the more general possibility of multiple flux calibrators.
Apply the Calibrations
In AIPS, the task CLCAL applies calibration solutions to a calibration table. In CASA the equivalent task is applycal. We loop over sources and calibrators to properly match them:
for source, calibrator in calDict.iteritems():
applycal(vis='ag733.ms', field=source,
gaintable='cal.Gflx', gainfield=calibrator,
gaincurve=False)
Baseline-based corrections
Although it makes only modest improvements for the present example, observations including a mix of EVLA and VLA telescopes may benefit from performing baseline-based corrections on a strong calibrator. Now that we have initial antenna-based solutions, we can use blcal.
default('blcal')
vis = 'ag733.ms'
# output baseline-based calibration solutions
caltable = 'cal.BL'
# use the strong flux calibrator to determine baseline-based solutions
field = '0137+331'
# generate a solution for each calibrator scan
solint = 'inf'
gaintable = 'cal.Gflx'
# calibration table with the best antenna-based solutions, so far
gainfield = '0137+331'
# calibrator for the BL calibrator; in this example, gain cal and bl cal are the same
interp = 'nearest'
gaincurve = False
# do the first day's data
timerange = '2006/12/09/0:0:0~24:0:0'
blcal()
# now the second day
timerange = '2006/12/13/0:0:0~24:0:0'
blcal()
This operation generates the calibration table cal.BL. Solutions can be inspected with plotcal. Now we have to use applycal again to apply both the antenna-based and baseline-based calibration solutions to the data.
for source, calibrator in calDict.iteritems():
applycal(vis='ag733.ms', field=source,
gaintable=['cal.Gflx','cal.BL'], gainfield=calibrator,
gaincurve=False)
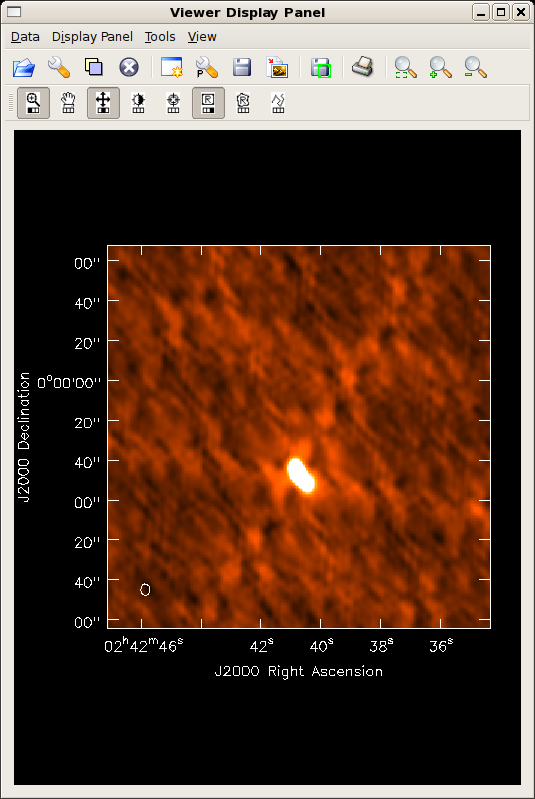
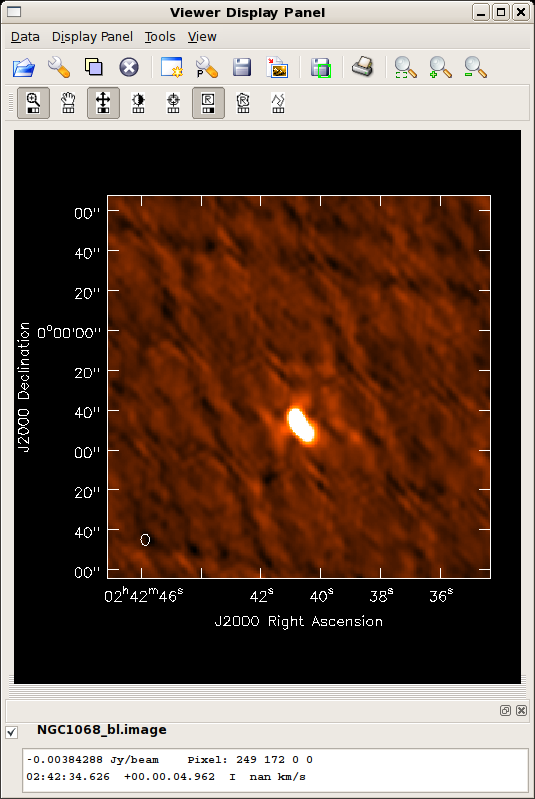
Did the blcal operation make a difference here? Getting ahead of ourselves a little, the plots at right show the initial clean images of NGC1068, one with blcal applied, and one without, and both images are displayed with the same stretch. There does appear a slight reduction of artifacts with blcal applied, but of course your mileage will vary.
Splitting the calibrated source data from the multisource measurement set
To split the calibrated source data out of the measurement set, use the CASA task split.
splitfile = 'NGC1667.split.ms'
rmtables(splitfile) # get rid of any old versions before splitting
split(vis='ag733.ms',outputvis=splitfile, datacolumn='corrected', field='NGC1667')
Alternatively, you can take advantage of scripting to loop over the sources.
for source in sourcelist:
splitfile = source + '.split.ms'
rmtables(splitfile)
split(vis='ag733.ms',outputvis=splitfile,
datacolumn='corrected', field=source)
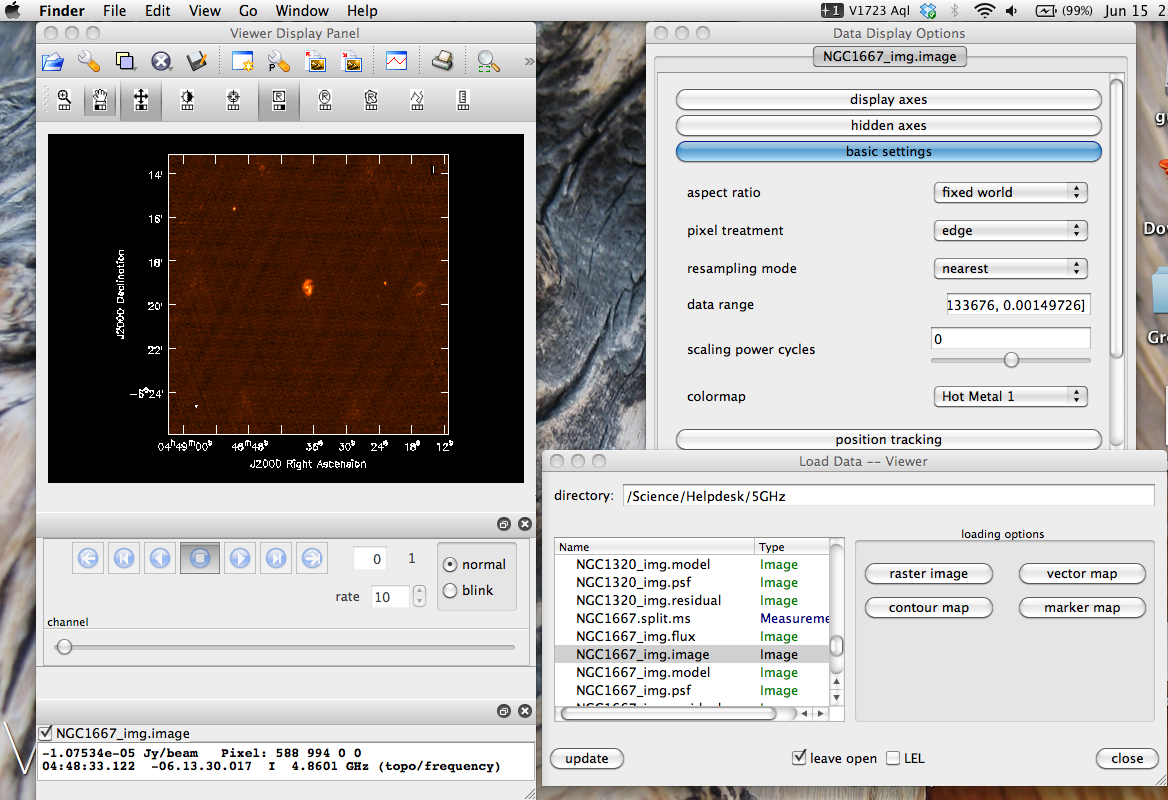
Imaging
Use clean for imaging and clean deconvolution. Like IMAGR in AIPS, it's worth some time mulling over all of the options. A complete list of keywords is given in the example, below. Most of the options should be relatively familiar to the AIPS user.
For this snapshot survey, the example chooses a fairly conservative, slow clean to accommodate the less-than-ideal beam. You could probably get away with more efficient cleaning, but it's better to play it safe in scripted self-calibration.
default(clean)
mode = 'mfs'
niter = 2000
gain = 0.1
threshold = '4.5e-5Jy'
psfmode = 'clark'
imagermode = 'csclean'
cyclefactor = 3
cyclespeedup = -1
multiscale = []
interactive = False
mask = []
imsize = [1024, 1024]
cell = ['0.75arcsec', '0.75arcsec']
stokes = 'I'
weighting = 'natural'
uvtaper = False
pbcor = False
minpb = 0.1
usescratch = False
async = False
for source in sourcelist:
splitfile = source + '.split.ms'
vis = splitfile
imagename = source + '_img'
rmtables(imagename + '*') # get rid of earlier versions of the image
clean()
Take a look at the image using viewer.
viewer()
From the GUI, select the .image file you want to view, select Raster Image, and then dismiss and close your way through to the display.
Self-calibration
At this point we have a set of calibrated visibilities (the "splits") and a set of images. There's not much more that can be done for the fainter sources, but the brighter sources will almost certainly show some residual calibration artifacts that can be cleaned up with self-calibration. The strategy is to repeat using the primary calibration application gaincal, only this time we use the source itself as a calibrator.
NGC 1068 is a good example of a difficult source. Firstly, it's very bright, so residual phase calibration errors should be obvious. Secondly, like the larger fraction of the sky, it's located painfully close to the celestial equator, meaning that the (u,v) coverage is particularly poor.
To view the calibration artifacts, use viewer to display the image of NGC 1068, but adjust the display levels so that the displayed image saturates at 10 mJy. To do this, select the leftmost button bearing a wrench icon. The "Data Display Options" GUI opens, and you can enter the desired display stretch in the Data Range text box. To cutoff the stretch at 30 mJy,
- Data Range: [-0.005,0.03]
You can see some residual systematics in the background and sidelobe artifacts that suggest the phase calibration isn't as good as it might be.
The process of self-calibration will involve iterating between (i) improved clean models of the source and (i+1) improved calibrations generated by gaincal. For simplicity, in this example we will repeatedly generate new calibration tables so that we can avoid using accum.
Step i
In principle, we could start with the initial clean model that we generated above. For this example, we'll take a more cautious approach and re-run clean interactively.
source = 'NGC1068' # set up a global variable to store the source we're working on
tget clean
vis = source + '.split.ms'
imagename = source + '_img0'
interactive = True
npercycle = 10
go
The trick to interactive cleaning is to draw clean boxes around the sources as they appear in the residual image. By default, clean boxes are set by right-clicking on the image and dragging. The box can be edited until you double right-click inside the box, which then sets that box for cleaning. Press the green icon to continue cleaning the image; on the next major cycle, a new residual map will be displayed, affording the option to add clean boxes as new sources appear where they were once swamped by sidelobes.
Of course, for self-calibration, it pays to stop the cleaning early when artifacts start to appear in the residuals!
Step i + 1
To start, we should already have a clean model for NGC 1068 stored in the visibility database. Now we can set up gaincal.
default('gaincal') # reset inputs for gaincal (akin to RESTORE in AIPS)
vis = source + '.split.ms'
caltable = source + '.gcal0'
gaintype = 'G'
calmode = 'p' # phase-only self-calibration at this stage
solint = '10 min' # set up a fairly coarse correction interval in this early stage
minsnr = 3.0
gaintable = ''
field = source
gainfield = source # are these necessary?
gaincurve = False
gaincal() # or go!
Pause for Reflection and Troubleshooting
Self-calibration is a method to fine-tune your original calibration, and, as such, should produce relatively small phase corrections assuming the primary calibration was applied correctly. Now is a good time to run plotcal on your new calibration table (NGC1068.gcal0) to make sure the phase corrections range from a few to perhaps 10 or 20 degrees. If you see phase corrections greatly exceeding 20 degrees, then there's a chance of either
- the primary calibration wasn't applied to the split database correctly, and should be checked, or
plotcal(caltable, iteration='antenna', xaxis='time', yaxis='phase', plotrange=[-1,-1,-180,180])
Step i + 2
Assuming plotcal gave reasonable results, next apply the calibration.
default('applycal')
vis = source + '.split.ms'
gaintable = source + '.gcal0'
gainfield = source
go
And now we can generate a new clean model.
tget clean
imagename=source + '_img1' # choose an image root name that matches the future cal table to facilitate bookkeeping
go
Further Steps
Assuming things are going well, we can now continue to repeat the process until we are satisfied with the image.
tget gaincal
caltable = source +'.gcal1'
solint = "5 min" # halve the solution interval.
go
# good idea to plotcal() again here!
tget applycal
gaintable = source + '.gcal1'
go
tget clean
imagename = source + '_img2'
go
And so on, decreasing the solution interval as much as the S/N and sampling allow.
Note that gaincal always uses the 'data' column (not the recently self-calibrated 'corrected' column) to find gain solutions. Therefore, these iterations simply serve to improve the data model, and gain solutions at each iteration are independent (e.g., the solution from the second self calibration iteration does not accumulate on top of the solution from the first interval. Use the CASA task accum if you wish to accumulate gain solutions).
One Last Iteration: Amplitude & Phase Self Calibration
After you have improved the phase calibration as much as possible, one last iteration where both phase and amplitude are calibrated can be beneficial. A&P self calibration should be performed with long solution intervals---typically the length of a scan.
default('gaincal') # reset inputs for gaincal (akin to RESTORE in AIPS)
vis = source + '.split.ms'
caltable = source + '.gcalap'
gaintype = 'G'
calmode = 'ap' # it's finally time to calibrate amplitudes, too
solint = 'inf' # one solution interval per scan (in conjunction with combine='')
combine = ''
minsnr = 3.0
gaintable = source + '.gcal1' # apply your best phase calibration on the fly.
gaincal() # or go!
Here, we are applying the best phase calibration solution on the fly by setting the gaintable parameter, so that this A&P self calibration iteration will only make small tweaks on top of the phase solutions we just worked so hard to find. Check your solutions in plotcal---amplitudes should not vary much and should be close to unity. Phase tweaks should be minimal and close to zero. If the solutions look reasonable, go ahead and apply the calibration:
default('applycal')
vis = source + '.split.ms'
gaintable = [source + '.gcal1', source + '.gcalap']
gainfield = source
go
The critical difference here from phase-only self calibration iterations is that we are applying two gain tables---the best phase solutions (source + '.gcal1') and the A&P solutions (source + '.gcalap'). Now you should be ready for one final imaging run with clean! For consistency, we will run it with the same command we used the first time:
default(clean)
mode = 'mfs'
niter = 2000
gain = 0.1
threshold = '4.5e-5Jy'
psfmode = 'clark'
imagermode = 'csclean'
cyclefactor = 3
cyclespeedup = -1
multiscale = []
interactive = False
mask = []
imsize = [1024, 1024]
cell = ['0.75arcsec', '0.75arcsec']
stokes = 'I'
weighting = 'natural'
uvtaper = False
pbcor = False
minpb = 0.1
async = False
usescratch = False
vis = 'NGC1068.split.ms'
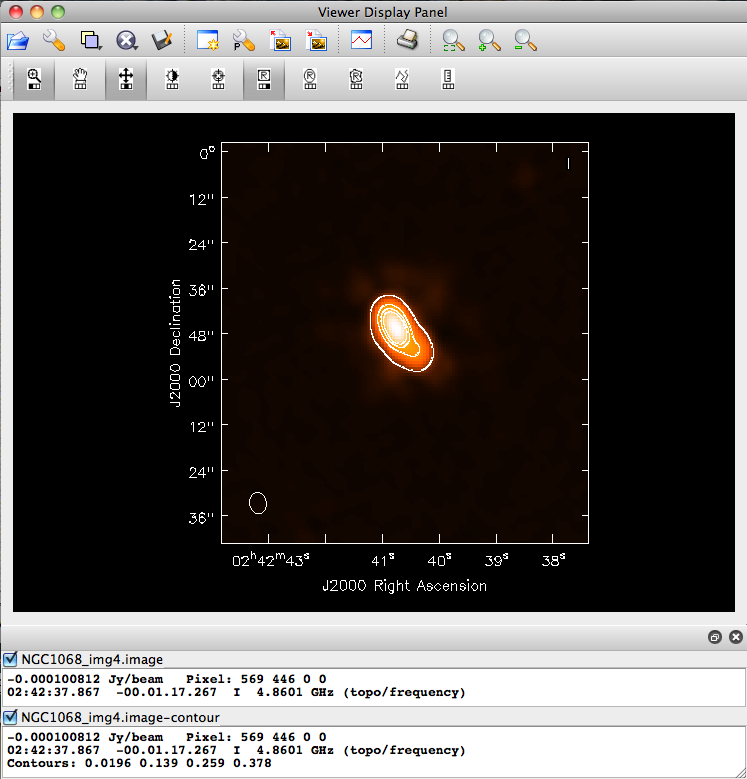
imagename = 'NGC1068_img4'
rmtables(imagename + '*') # get rid of earlier versions of the image
clean()
Last checked on CASA Version 3.4.0.