Imaging Prep
Imaging Prep Workflow
[File:Imaging_Prep_Flow.png]
Get a list of ms files to image
<figure id="Weblog_Home.png">
</figure> Once the calibration has run, it will produce ‘*.ms.split.cal’ files for each execution. These files contain the calibrated data. Run the following commands to grab the names of all files in the directory that have the extension ‘.ms.split.cal’ and place them into a list called vislist.
# in CASA
import glob
vislist=glob.glob('*.ms.split.cal')
Review the Calibration
Once the list of measurement set (ms) files have been collected, review the calibration to make sure it appears reasonable. The results of the calibration are in the qa directory of your delivered package. If the dataset is pipeline calibrated, download and untar the pipeline file in the qa directory by using <figure id="Weblog_MS_Overview.png">
</figure>
# in bash
for i in $(ls *.tar); do tar -xvf $i; done
# or in CASA
For i in glob.glob(‘*.tar’):
os.system(‘tar -xvf %s’ % (i))
Once this is done, look within the /html directory for the index.html file. This contains the html code for displaying the contents of the qa directory in a web browser. The command below opens the main page of the weblog in firefox.
firefox index.html
<figure id="Weblog_By_Task.png">
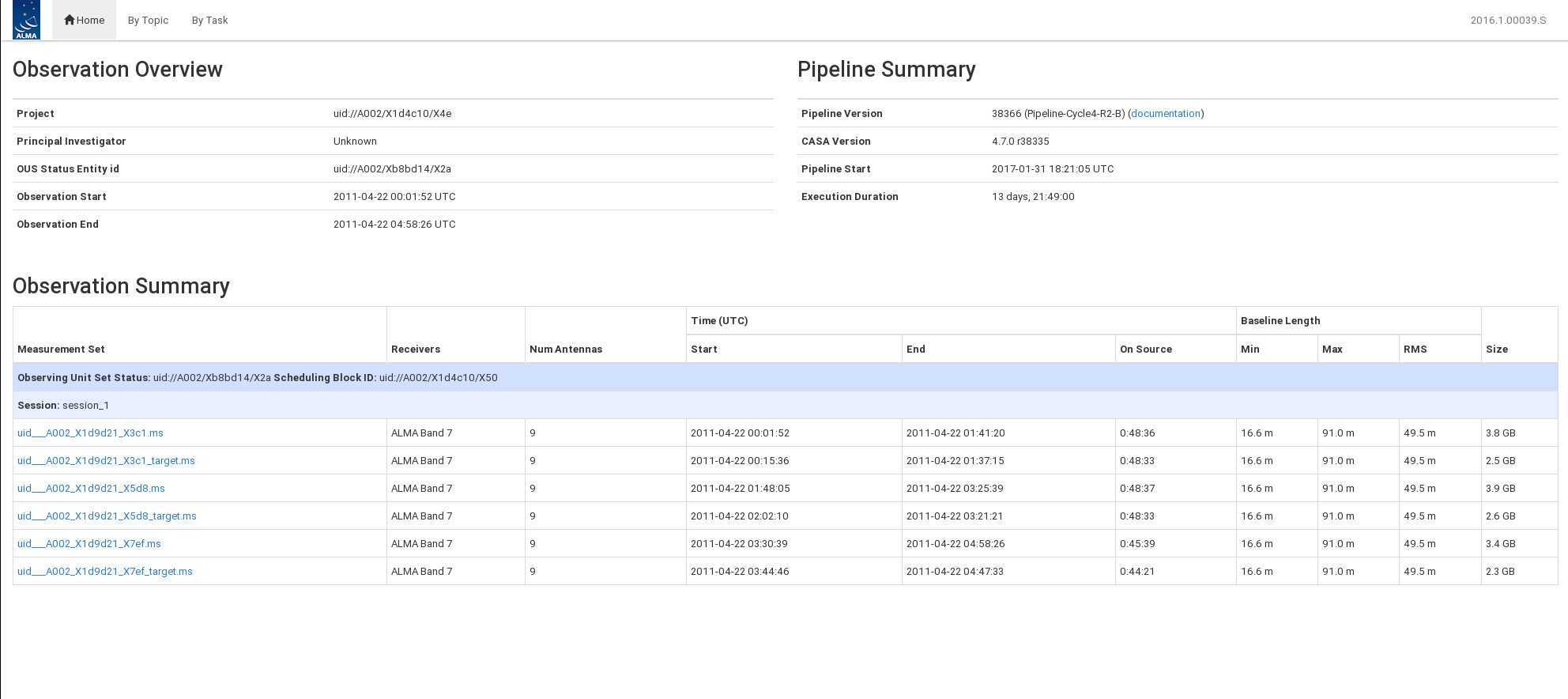
</figure> The standard opening page has three main sections: observation overview, pipeline summary, and observation summary. This is shown in Figure 1.
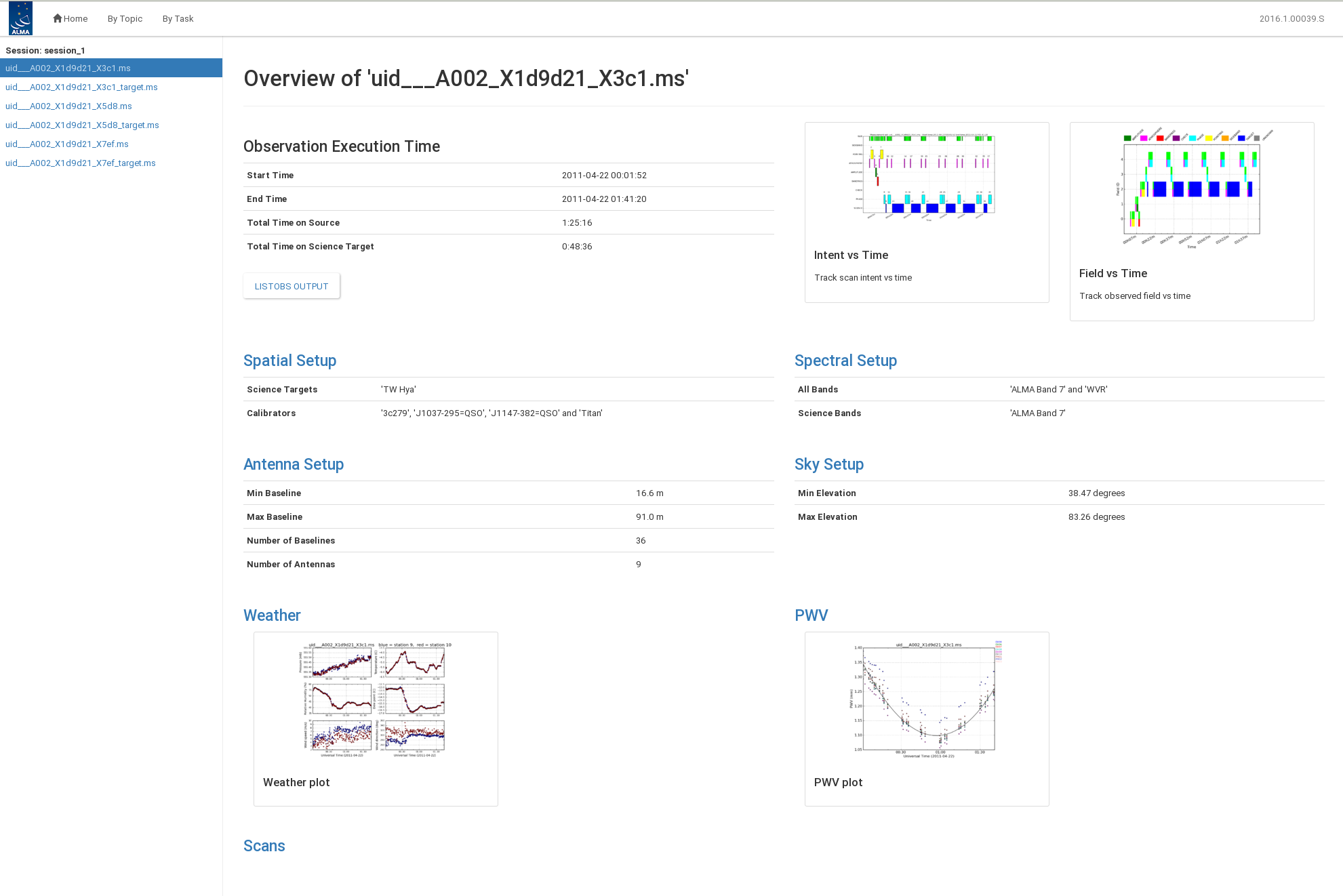
From the opening page, you can navigate to view the calibration By Topic and By Task and open observation specific information by clicking on the ms name. Figure 2 shows an example of an ms summary page.
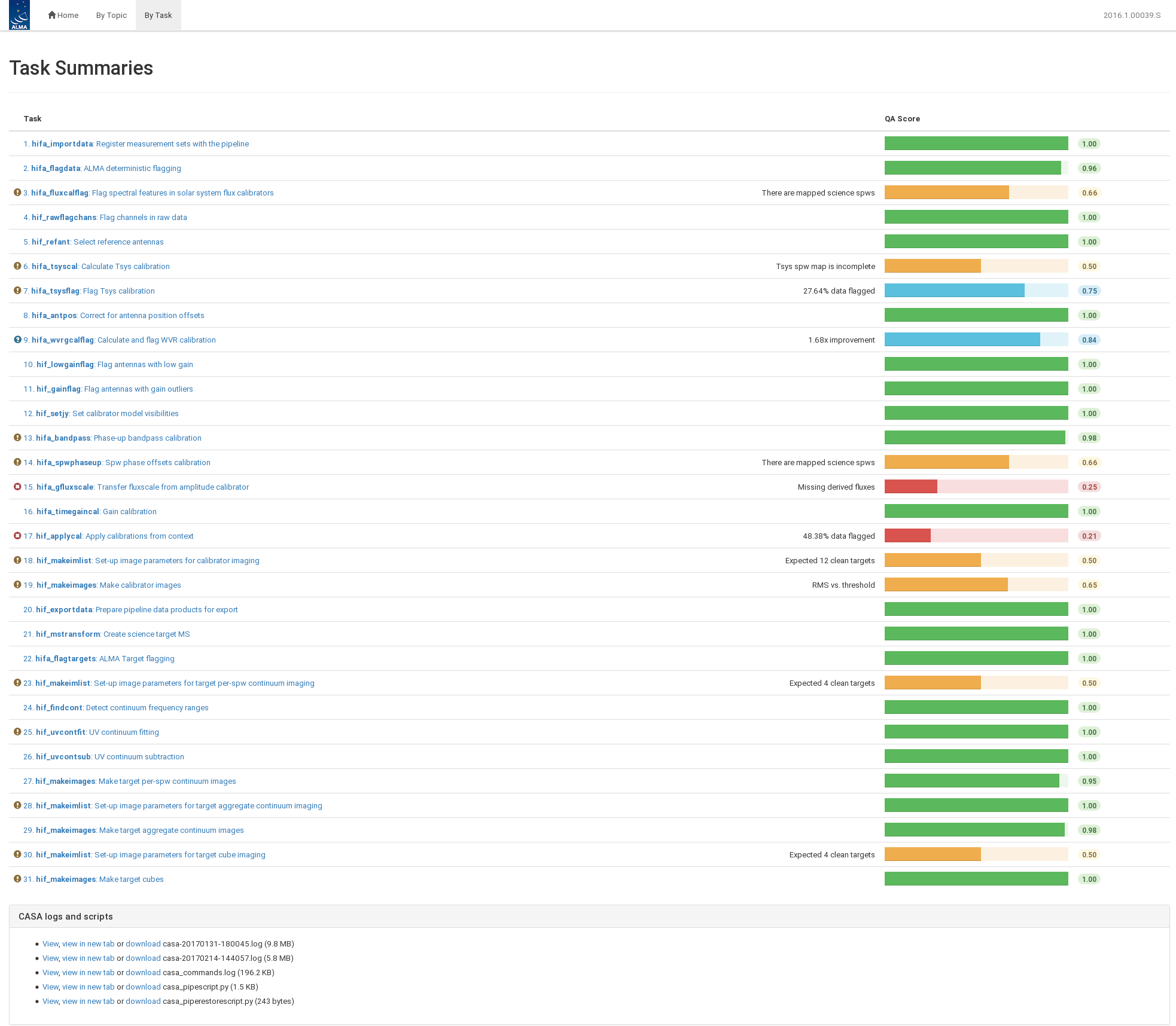
Figure 3 shows the By Task view. Each task can be opened to view important calibration plots, tables, and flagging statistics.
If the data is manually calibrated, you can review the calibration with the *.png and *.txt files in the qa directory. These display helpful plots and statistics about the calibration.
Flag bad data (optional)
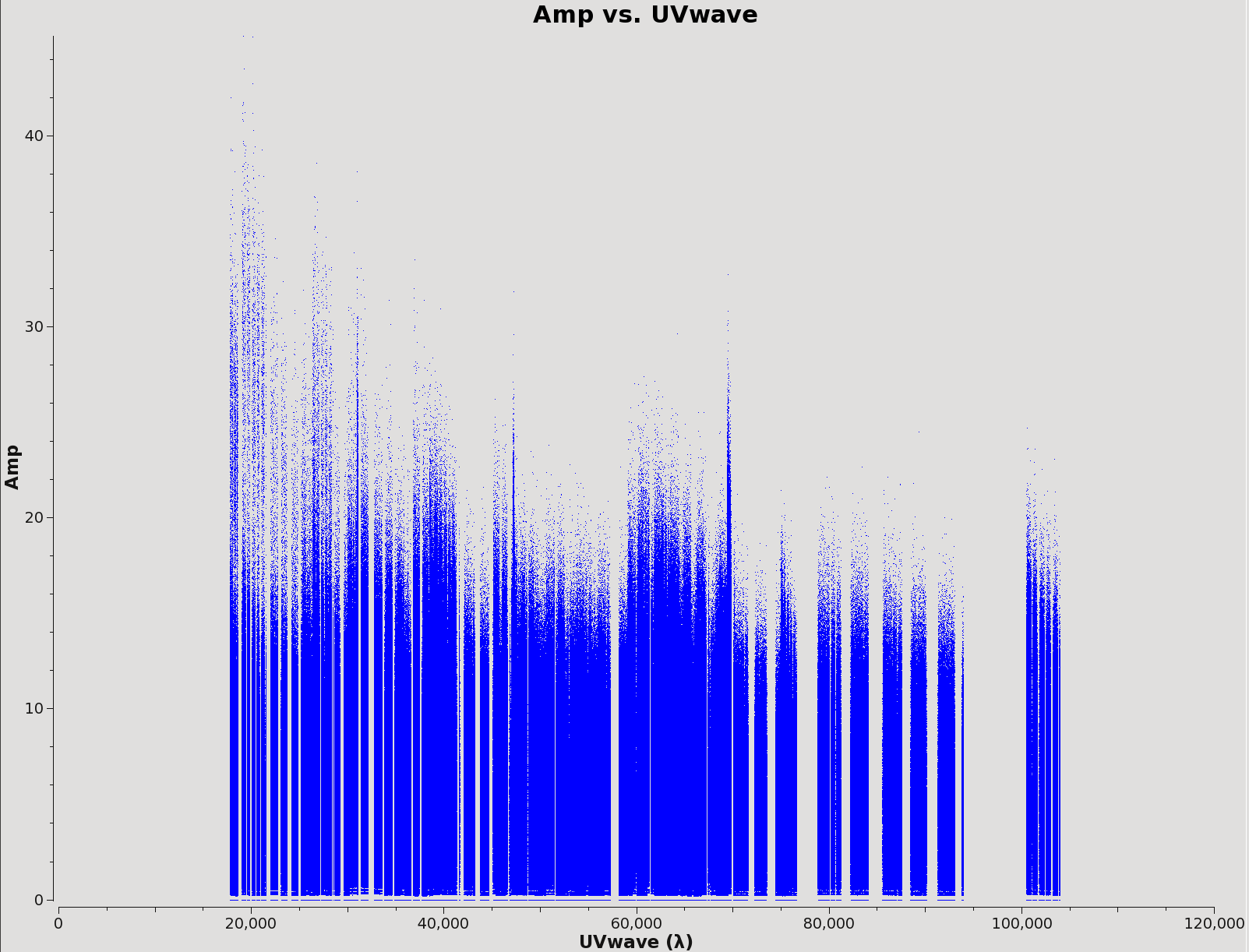
plotms should be used to find worrisome data points by iterating through fields and spws. This should be done on the *.ms.split.cal files in the calibrated directory. The remainder of this guide should be run in CASA. Figure 4 shows an example of TW Hydra science spw 2
# in CASA
fieldlist = ['3'] # change to list of science data fields to inspect
spwlist = ['1'] # change to list of science spws to inspect
for vis in vislist:
for field in fieldlist:
for spw in spwlist:
plotms(vis=vis,xaxis='uvwave',yaxis='amp',avgtime='3e8',
field=field,spw=spw)
raw_input("push enter to continue")
plotms(vis=vis,xaxis='chan',yaxis='amp',avgtime='3e8',
field=field,spw=spw)
raw_input("push enter to continue")
Save the original flags of each ms prior to flagging any additional data.
# in CASA
for vis in vislist:
flagmanager(vis=vis,
mode='save',
versionname='original_flags')
Use flagdata if you find any additional antennas, data, SPW, etc which need to be flagged. You can use the locate buttons in plotms and plotcal to identify what needs to be flagged. Update the following command with the specific ms name with the vis parameter and any antenna, SPW, etc. Once the data is flagged, the flagging can be checked by using plotms. <figure id="TWHydra_corrected_Spw2.png">
</figure>
# in CASA
flagdata(vis='',mode='manual',action='apply',flagbackup=False)
If you need to restore original flags, use the following command. You will need to update the ms name in the vis parameter.
# in CASA
flagmanager(vis='',mode='restore',versionname='original_flags')
Flux Equalization (optional)
Flux equalization can be used if you have multiple executions using the same phase calibrator. <figure id="Gfluxscale table.png">
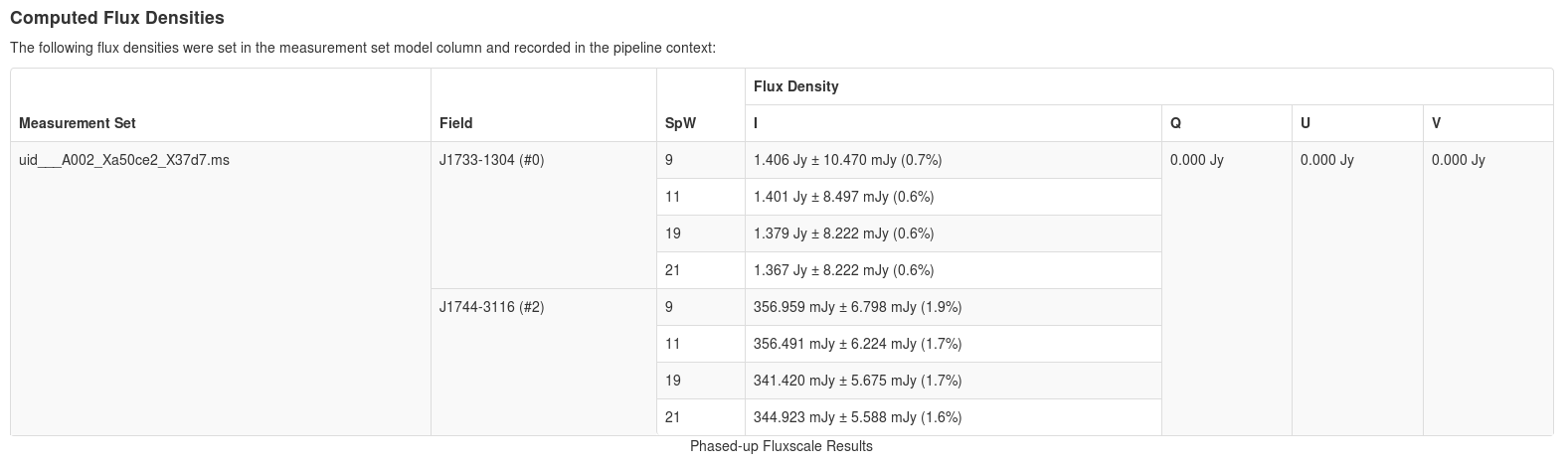
</figure> If you have determined flux equalization is required for your data, please file a Helpdesk ticket (help.almascience.org) and a NAASC staff member can assist you by generating a script for your measurement sets. Flux equalization is not generally recommended unless deemed absolutely necessary. ALMA Memo 434 outlines various uncertainties relevant to amplitude calibraion . You can refer to the respective cycle’s Proposer’s Guide (Cycle 4) for expected uncertainties. In addition, the ALMA Calibrator Source Catalog can be used to compare variations detected from source monitoring.
If your dataset was pipeline calibrated, the derived flux densities of your calibrators can be found in the hifa_gfluxscale step of the weblog. If your dataset was manually calibrated, the *.fluxscale file of each execution contains derived flux densities.
The first step in flux equalization is individually setting the flux density of the phase calibrator in each spectral window using setjy. Then amplitude calibration tables are generated and applied using gaincal and applycal respectively. Finally, all calibrated measurement sets will be concatenated using concat.
If you have performed flux equalization, you can skip to the Imaging_Prep#Splitting off science target data section below.
Combining measurement sets from multiple executions
This section will either need to be commented out or deleted if you have only one execution. However, if there are multiple *.ms.split.cal files within the calibrated directory, you will want to combine them all into one single ms file in order to make imaging more manageable later on.
Each execution of the scheduling block will generate multiple spectral windows with different sky frequencies, but the same rest frequency, due to the motion of the Earth. Thus, the resulting concatenated file will contain n spws, where n is (number of original science spws) x (number of executions). In other words, the multiple spws associated with a single rest frequency will not be regridded to a single spectral window in the ms. Combine all executions into one ms file, called calibrated.ms, using the concat command.
# in CASA
concatvis='calibrated.ms'
rmtables(concatvis)
os.system('rm -rf ' + concatvis + '.flagversions')
concat(vis=vislist,
concatvis=concatvis)
Splitting off science target data
The calibrated.ms produced in section IV, or your original ms file if you only have one execution, will not only contain the science target(s), but also all calibrator sources observed. At this point, you are only concerned with imaging our science targets so you can split off all non science fields from the ms. This will reduce the size of the file and make it more manageable during imaging. Depending on your project there may be several executions or only one. Uncomment the line that fits with your data to set the concatvis variable.
# in CASA
# Uncomment following line for single executions
# concatvis = vislist[0]
# Uncomment following line for multiple executions
# concatvis='calibrated.ms'
You will split out all target fields from the file and copy them, with only the data column, into a new ms file called calibrated_source.ms. Follow this link for more information regarding the structure of measurement sets.
# in CASA
sourcevis='calibrated_source.ms'
rmtables(sourcevis)
os.system('rm -rf ' + sourcevis + '.flagversions')
split(vis=concatvis,
intent='*TARGET*', # split off the target sources
outputvis=sourcevis,
datacolumn='data')
At this point, it is convenient to create a listobs file to reference during imaging. Inspect the listobs to make sure the split and concat worked as desired.
# in CASA
listobs(vis=sourcevis,listfile=sourcevis+'.listobs.txt')
An example listobs file is below.
================================================================================
Observer: Unknown Project: T.B.D.
Observation: ALMA
Telescope Observation Date Observer Project
ALMA [ 4.81015e+09, 4.81015e+09]Unknown T.B.D.
ALMA [ 4.81015e+09, 4.81016e+09]Unknown T.B.D.
ALMA [ 4.81016e+09, 4.81017e+09]Unknown T.B.D.
Data records: 126900 Total elapsed time = 16902.1 seconds
Observed from 22-Apr-2011/00:15:36.7 to 22-Apr-2011/04:57:18.8 (UTC)
ObservationID = 0 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent
22-Apr-2011/00:15:36.7 - 00:16:07.0 9 0 TW Hya 540 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
00:20:57.8 - 00:30:43.2 13 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
00:36:49.2 - 00:44:43.5 18 0 TW Hya 7020 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
00:49:33.9 - 00:59:19.3 22 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
01:05:46.8 - 01:13:41.2 27 0 TW Hya 7020 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
01:20:07.2 - 01:29:52.6 31 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
01:37:15.6 - 01:40:13.9 36 0 TW Hya 2700 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
ObservationID = 1 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent
22-Apr-2011/02:02:10.9 - 02:02:41.1 44 0 TW Hya 540 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
02:07:36.7 - 02:17:22.0 48 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
02:23:35.7 - 02:31:30.0 53 0 TW Hya 7020 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
02:36:23.6 - 02:46:09.0 57 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
02:52:40.2 - 03:00:34.6 62 0 TW Hya 7020 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
03:05:14.0 - 03:14:59.4 66 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
03:21:21.4 - 03:24:19.6 71 0 TW Hya 2700 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
ObservationID = 2 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent
22-Apr-2011/03:44:46.9 - 03:45:17.1 79 0 TW Hya 540 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
03:50:08.4 - 03:59:53.7 83 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
04:06:03.7 - 04:13:58.1 88 0 TW Hya 7020 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
04:18:49.1 - 04:28:34.4 92 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
04:34:59.5 - 04:42:53.9 97 0 TW Hya 7020 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
04:47:33.5 - 04:57:18.8 101 0 TW Hya 8640 [0,1,2,3] [10.1, 10.1, 10.1, 10.1] [OBSERVE_TARGET#ON_SOURCE]
(nRows = Total number of rows per scan)
Fields: 1
ID Code Name RA Decl Epoch SrcId nRows
0 none TW Hya 11:01:51.844983 -34.42.17.16088 J2000 0 126900
Spectral Windows: (4 unique spectral windows and 1 unique polarization setups)
SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) Corrs
0 3840 TOPO 356497.936 122.070 468750.0 356732.2500 XX YY
1 3840 TOPO 357734.314 122.070 468750.0 357968.6279 XX YY
2 3840 TOPO 346034.314 122.070 468750.0 345800.0000 XX YY
3 3840 TOPO 343955.936 122.070 468750.0 343721.6221 XX YY
Sources: 4
ID Name SpwId RestFreq(MHz) SysVel(km/s)
0 TW Hya 0 - -
0 TW Hya 1 - -
0 TW Hya 2 - -
0 TW Hya 3 - -
Antennas: 9:
ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m)
East North Elevation x y z
0 DV04 J505 12.0 m -067.45.18.0 -22.53.22.8 -7.2141 -541.3485 15.0178 2225061.036842 -5440128.036234 -2481534.422455
1 DV06 T704 12.0 m -067.45.16.2 -22.53.22.1 42.8987 -520.1911 15.0694 2225110.551677 -5440116.726350 -2481514.951072
2 DV07 J510 12.0 m -067.45.17.8 -22.53.23.5 -0.3652 -563.8032 14.9605 2225064.049398 -5440117.310745 -2481555.086720
3 DV08 T703 12.0 m -067.45.16.2 -22.53.23.9 42.8798 -575.6928 14.6278 2225102.207484 -5440096.375809 -2481565.910698
4 DV09 N602 12.0 m -067.45.17.4 -22.53.22.3 8.8012 -527.8598 15.0513 2225077.857538 -5440126.858119 -2481522.008896
5 DV10 N606 12.0 m -067.45.17.1 -22.53.23.6 19.1981 -566.5667 14.9520 2225081.746122 -5440108.902762 -2481557.629365
6 PM01 T702 12.0 m -067.45.18.6 -22.53.24.1 -23.6269 -582.3103 14.9195 2225039.780230 -5440119.418780 -2481572.120530
7 PM02 T701 12.0 m -067.45.18.8 -22.53.22.2 -29.1268 -522.7917 15.0566 2225043.501617 -5440143.045000 -2481517.341976
8 PM03 J504 12.0 m -067.45.17.0 -22.53.23.0 22.2015 -550.2548 14.9948 2225086.942689 -5440113.674706 -2481542.618537
Regridding spectral window (optional)
The NA imaging team strongly recommends using the clean command to regrid spectral windows on the fly. An exception to this recommendation are cases where lines shift too much between executions to identify a common channel range for continuum subtraction. Regridding can be used when there are multiple executions done at different times for the same target, an extremely high redshifted project, ephemeris projects, or when spws or images have different coordinate systems. In general, if the rest frame between different spws/images needs to be used, regridding using the cvel command as opposed to just using the clean command could be optimal for the dataset. If this is not necessary for your project, proceed to the next section.
Due to a bug in all released versions of CASA, mstransform and cvel should not be used to average greater than 2 channels together. This averaging should be done using clean until the bug is resolved.
We will start by setting the parameters specific to your observations.
# in CASA
sourcevis='calibrated_source.ms'
regridvis='calibrated_source_regrid.ms'
veltype = 'radio' # Keep set to radio. See notes in imaging section.
width = '0.23km/s' # due to bug in cvel/mstransform, do not regrid > 2 channels
nchan = -1 # leave this as the default
mode='velocity' # see science goals in the OT
start='' # leave this as the default
outframe = 'bary' # velocity reference frame. see science goals in the OT.
restfreq='115.27120GHz' # rest frequency of primary line of interest.
field = '4' # select science fields.
spw = '0,5,10' # spws associated with a single rest frequency. Do not attempt to combine spectral windows associated with different rest frequencies. This will take a long time to regrid and most likely isn't what you want.
Regridding here uses the cvel command to regrid multiple spws associated with a single rest frequency into a single spw. To avoid clean regridding the image a second time you will need to use the same parameters used for cvel for clean.
# in CASA
rmtables(regridvis)
os.system('rm -rf ' + regridvis + '.flagversions')
cvel(vis=sourcevis,
field=field,
outputvis=regridvis,
spw=spw,
mode=mode,
nchan=nchan,
width=width,
start=start,
restfreq=restfreq,
outframe=outframe,
veltype=veltype)
Rename and backup data set
Depending on the different tasks that were performed above, the current measurement set will have a variety of names. For ease of beginning imaging with a common measurement set, rename the file to calibrated_final.ms.
If you haven’t regridded, your current ms should be named calibrated_source.ms.
# in CASA
# uncomment if you haven’t regridded
# os.system('mv -i ' + sourcevis + ' ' + 'calibrated_final.ms')
If you have regridded, your current ms should be named calibrated_source_regrid.ms.
# in CASA
# uncomment if you have regridded
# os.system('mv -i ' + regridvis + ' ' + 'calibrated_final.ms')
Now that you have one fully calibrated measurement set to image, we recommend creating a backup of the dataset. It is easy to corrupt a measurement set during clean, tclean, and uvcontsub so it is a good idea to have a backup dataset.
# in CASA
os.system('cp -ir calibrated_final.ms calibrated_final.ms.backup')
Continue to Imaging
This guide is continued at Image_Continuum