AntennaeBand7 North Calibration 3.4
- This script assumes that you have downloaded Antennae_Band7_UnCalibratedMSAndTablesForReduction.tgz from AntennaeBand7#Obtaining_the_Data.
- Details of the ALMA observations are provided at AntennaeBand7_3.4.
- This casaguide covers the processing of a subset of the data for the Northern mosaic.
- This portion of the guide covers calibration of the raw visibility data. To skip to the imaging portion of the guide, see: AntennaeBand7 North Imaging 3.4.
Overview
This part of the Antennae Band 7 CASA guide will step you through the calibration of the visibility data for the Northern mosaic. We will begin by flagging (marking as bad) data known to be useless before any inspection, for example data where one telescope blocks the line of sight of another. Then we will apply telescope-generated calibration tables to partially correct for atmospheric effects. After inspecting the data, we will flag some additional data that exhibit pathologies. Then we will use observations of the calibrators Titan and 3c279 to derive the phase and amplitude response of individual antennas as a function of time and frequency ("phase", "amplitude", and "bandpass" calibrations). We will apply these to the data and then extract the calibrated source data into a file appropriate for imaging.
The general procedure in this guide follows the other ALMA CASA guides: NGC3256Band3 and TWHydraBand7_3.4.
Unpack the Data
Once you have downloaded the Antennae_Band7_UnCalibratedMSandTablesForReduction.tgz, unpack the file in a terminal outside CASA using
tar -xvzf Antennae_Band7_UnCalibratedMSandTablesForReduction.tgz
then change directory to the new directory (created by the unpacking)
cd Antennae_Band7_UnCalibratedMSandTablesForReduction
You may wish to type
ls
to look at the files present. You should see a set of files with extension ".ms". These are CASA measurement set (MS) files. The data have already been converted to MS format from the native ALMA format using the CASA task importasdm. In addition to the data, we provide you with calibration tables containing system temperature (Tsys), water vapor radiometer (WVR), and antenna position information. For Early Science, these tables will either be pre-applied or supplied with the data.
This guide requires Python module analysisUtils. If you have not already installed analysisUtils please follow the link to do so.
To begin, start CASA by typing
casapy
Be sure that you are using the version indicated at the top of this page.
Confirm your version of CASA
This guide has been written for CASA release 3.4.0. Please confirm your version before proceeding.
# In CASA
version = casalog.version()
print "You are using " + version
if (int(version.split()[4][1:-1]) < 19407):
print "\033[91m YOUR VERSION OF CASA IS TOO OLD FOR THIS GUIDE."
print "\033[91m PLEASE UPDATE IT BEFORE PROCEEDING."
else:
print "Your version of CASA is appropriate for this guide."
Install Analysis Utilities
Analysis Utilities (or analysisUtils for short) is a small set of Python scripts that provide a number of analysis and plotting utilities for ALMA data reduction. This guide uses a few of these utilities. They are very easy to install (just download and untar). See
http://casaguides.nrao.edu/index.php?title=Analysis_Utilities
for a full description and download instructions. If you do not wish to do this, see the CASA 3.3 version of the guide linked at the top of this page for alternative (but slow) plotting options. Analysis Utilities are updated frequently so if its been a while since you installed it, its probably worth doing it again. If you are at an ALMA site or ARC, the analysis utilities are probably already installed and up to date.
Initial Inspection
First we will take stock of what we have. If you have not already done so, begin by reviewing the description of the observations here: AntennaeBand7_3.4. In this tutorial we only deal with the 4 data sets for the Northern mosaic:
Northern Mosaic:
- uid___A002_X1ff7b0_Xb.ms
- uid___A002_X207fe4_X3a.ms
- uid___A002_X207fe4_X3b9.ms
- uid___A002_X2181fb_X49.ms
The first step is to get basic information about the data: targets observed, time range, spectral setup, and so on. We do this using the task listobs, which will output a detailed summary of each dataset. Enter the following commands into CASA:
# In CASA
# Define a python list holding the names of all of our data sets
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49"]
# Loop over each element in the list and create summary file using listobs
for asdm in basename_north:
os.system('rm '+asdm+'.listobs.txt')
listobs(vis=asdm+'.ms', listfile=asdm+'.listobs.txt', verbose=True)
Note that after cutting and pasting a 'for' loop like this you often have to press return twice to execute. You may also want to take care to paste a line at a time if you are having trouble copy and pasting. Even better, you can use "cpaste" to paste blocks of code. To do so type "cpaste" at the CASA prompt, paste your commands, and then type "--" and hit return on the final (otherwise empty) line. This should look something like this:
CASA <8>: cpaste Pasting code; enter '--' alone on the line to stop. :basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9", : "uid___A002_X2181fb_X49"] : :for asdm in basename_north: : print asdm :-- uid___A002_X1ff7b0_Xb uid___A002_X207fe4_X3a uid___A002_X207fe4_X3b9 uid___A002_X2181fb_X49 CASA <9>:
cpaste should be much more robust than copying-and-pasting directly into the shell but if you have trouble, just carefully paste one line at a time directly into CASA and hit return until the desired command executes.
These commands define a python list called "basename_north", which contains the name of all 4 MS files for the North. The "for" loop executes for each element in basename_all, calling listobs and directing the output to a file called, e.g., "uid___A002_X1ff7b0_Xb.listobs.txt" for the first measurement set. You can browse through the listobs output as you would normally look at a text file (use emacs, vi, or another editor). You can also send the output to the terminal from inside of CASA. To do so type:
# In CASA
os.system('cat uid___A002_X1ff7b0_Xb.listobs.txt')
or
# In CASA
os.system('more uid___A002_X1ff7b0_Xb.listobs.txt')
CASA knows a few basic shell commands like 'cat', 'ls', and 'rm' but for more complex commands you will need to run them inside 'os.system("command")'. For more information see http://casa.nrao.edu/ .
Here is an example of the (abridged) output from listobs for the first dataset in the list, uid___A002_X1ff7b0_Xb.ms, which targets the Northern Mosaic. You would see this if you had specified verbose to be False in the listobs call:
================================================================================
MeasurementSet Name: uid___A002_X1ff7b0_Xb.ms MS Version 2
================================================================================
Observer: Unknown Project: T.B.D.
Observation: ALMA(11 antennas)
Data records: 181357 Total integration time = 4931.71 seconds
Observed from 28-May-2011/01:25:27.6 to 28-May-2011/02:47:39.3 (UTC)
Fields: 26
ID Code Name RA Decl Epoch SrcId nRows
0 none 3c279 12:56:11.16657 -05.47.21.5247 J2000 0 39116
1 none Titan 12:42:43.94813 -01.43.38.3190 J2000 1 10626
2 none NGC4038 - Antennae *12:01:53.17008 -18.52.37.9200 J2000 2 4818
3 none NGC4038 - Antennae *12:01:51.90301 -18.51.49.9437 J2000 2 6435
4 none NGC4038 - Antennae *12:01:52.43086 -18.51.49.9437 J2000 2 6424
5 none NGC4038 - Antennae *12:01:52.95871 -18.51.49.9437 J2000 2 5478
6 none NGC4038 - Antennae *12:01:53.48656 -18.51.49.9436 J2000 2 6435
7 none NGC4038 - Antennae *12:01:54.01441 -18.51.49.9436 J2000 2 6446
8 none NGC4038 - Antennae *12:01:52.16693 -18.51.56.4319 J2000 2 6435
9 none NGC4038 - Antennae *12:01:52.69478 -18.51.56.4318 J2000 2 6424
10 none NGC4038 - Antennae *12:01:53.22263 -18.51.56.4318 J2000 2 6446
11 none NGC4038 - Antennae *12:01:53.75049 -18.51.56.4318 J2000 2 6446
12 none NGC4038 - Antennae *12:01:51.90301 -18.52.02.9201 J2000 2 6435
13 none NGC4038 - Antennae *12:01:52.43086 -18.52.02.9200 J2000 2 6424
14 none NGC4038 - Antennae *12:01:52.95871 -18.52.02.9200 J2000 2 4818
15 none NGC4038 - Antennae *12:01:53.48656 -18.52.02.9200 J2000 2 4829
16 none NGC4038 - Antennae *12:01:54.01441 -18.52.02.9199 J2000 2 4829
17 none NGC4038 - Antennae *12:01:52.16694 -18.52.09.4082 J2000 2 4829
18 none NGC4038 - Antennae *12:01:52.69479 -18.52.09.4082 J2000 2 4829
19 none NGC4038 - Antennae *12:01:53.22264 -18.52.09.4082 J2000 2 3872
20 none NGC4038 - Antennae *12:01:53.75049 -18.52.09.4081 J2000 2 4829
21 none NGC4038 - Antennae *12:01:51.90301 -18.52.15.8964 J2000 2 4829
22 none NGC4038 - Antennae *12:01:52.43087 -18.52.15.8964 J2000 2 4829
23 none NGC4038 - Antennae *12:01:52.95872 -18.52.15.8963 J2000 2 4818
24 none NGC4038 - Antennae *12:01:53.48657 -18.52.15.8963 J2000 2 4818
25 none NGC4038 - Antennae *12:01:54.01442 -18.52.15.8963 J2000 2 4840
Spectral Windows: (9 unique spectral windows and 2 unique polarization setups)
SpwID #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) Corrs
0 4 TOPO 184550 1500000 7500000 I
1 3840 TOPO 344845.586 488.28125 1875000 XX YY
2 1 TOPO 343908.086 1875000 1875000 XX YY
3 3840 TOPO 354971.074 488.28125 1875000 XX YY
4 1 TOPO 343908.086 1875000 1875000 XX YY
5 128 TOPO 344900.518 15625 2000000 XX YY
6 1 TOPO 343892.705 1796875 1796875 XX YY
7 128 TOPO 354916.143 15625 2000000 XX YY
8 1 TOPO 343892.705 1796875 1796875 XX YY
Antennas: 11 'name'='station'
ID= 0-3: 'DV02'='A015', 'DV04'='J505', 'DV06'='T704', 'DV07'='A004',
ID= 4-7: 'DV08'='A072', 'DV09'='A008', 'DV10'='A009', 'DV11'='A016',
ID= 8-10: 'PM01'='T702', 'PM02'='A017', 'PM03'='J504'
================================================================================
If you set verbose=True you get a scan listing in addition to this, plus Source table and expanded Antenna table information.
This output shows that three sources were observed in each data set: 3c279, Titan, and the Antennae.
- For the Northern Mosaic our target fields are named "NGC4038 - Antennae" where each field is an individual mosaic pointing. There are 23 for the Northern Mosaic.
- Titan is observed once and will be used to set the absolute flux scale of the data.
- 3c279 plays two roles: it will serve as our bandpass calibrator, to characterize the frequency response of the antennas, and because it is fairly close on the sky to the Antennae (18.6 degrees away) it will serve as our secondary calibrator (also referred to as the "phase calibrator" or "gain calibrator"), to track changes in the phase and amplitude response of the telescopes over time. Observations of 3c279 are interleaved with observations of the Antennae.
The output also shows that the data contain many spectral windows. Using the labeling scheme in the listobs above these are:
- spw 0 targets ~185 GHz and holds water vapor radiometer data
- spw 1 and spw 3 hold our science data. These are "Frequency Domain Mode" (FDM) data with small (0.49 MHz) channel width and wide (1.875 GHz) total bandwidth. As a result these have a lot of channels (3840). spw 1 holds the lower sideband (LSB) data and includes the CO(3-2) line. We will focus on these data. For the CO(3-2) line the channel width corresponds to 0.426 km/s and the bandwidth of spw 1 to 1634 km/s.
- spw 2 and spw 4 hold frequency-averaged versions of spw 1 and 3 ("Channel 0" for those familiar with AIPS). These are used for some quick automated inspection. We will not use them here but we will carry out an equivalent inspection using spw 1.
- spw 5 and spw 7 hold lower a resolution processing ("Time Domain Mode", TDM) of the data from the same part of the spectrum (baseband) as spws 1 and 3. These data have only 128 channels across 2 GHz bandwidth and so have a much coarser channel spacing than the FDM data. These were used to generate the calibration tables that we include in the tarball but will not otherwise appear in this guide.
The final column of the listobs output in the logger (not shown above) gives the scan intent. Later we will use this information to flag the pointing scans and the hot and ambient load calibration scans.
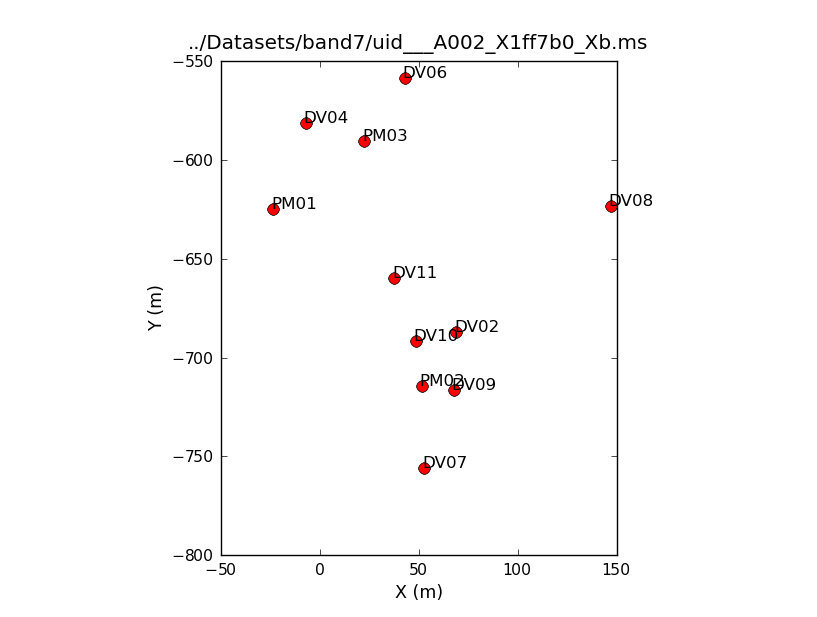
We'll now have a look at the configuration of the antennas used to take the data using the task plotants (<xr id="Uid_A002_X1ff7b0_Xb.ANT.png"/>).
<figure id="Uid_A002_X1ff7b0_Xb.ANT.png">
</figure>
# In CASA
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49"]
for asdm in basename_north:
print "Antenna configuration for : "+asdm
plotants(vis=asdm+'.ms', figfile=asdm+'.plotants.png')
clearstat()
dummy_string = raw_input("Hit <Enter> to see the antenna configuration for the next data set.")
This will loop through all the data sets, show you the antenna position for each, and save that as a file named, e.g., "uid___A002_X1ff7b0_Xb.plotants.png" for the first data set. The "raw_input" command asks CASA to wait for your input before proceeding. If you would prefer to just browse the .png files after the fact you can remove this. Notice that the antenna setup changes, but only slightly, over the course of the 10 data sets.
How to Deal With 4 Measurement Sets
It should already be clear from the initial inspection that dealing with 4 (or more) data sets at the same time can be a bit problematic. This is especially tricky in our case because the Antennae data contain two distinct sets of observations: the Northern and Southern Mosaics. The source name changes between these two scripts and there are different numbers of fields in the mosaic. We get around much of this issue by dealing with only the 4 Northern Mosaic datasets and ignoring the 6 Southern ones.
As a general rule one would reduce each individual observation separately or at the very least only group data observed in a uniform way and very close in time.
Unfortunately, a CASA Guide stepping through the reduction for each of 10 data sets would quickly become unwieldy. Therefore we will use a few tricks to reduce the Antennae data in a kind of batch mode. You have already seen the first trick: defined the python list variable basename_north holding the names of each data set so we can loop over this list to execute the same command on each data set.
You only need to define your list of MS files once per CASA session. Then basename_north will be a variable in the casapy shell. You can check if it exists by typing print basename_north. In the interests of allowing you to easily exit and restart CASA and pick this guide up at any point we will redefine basename_north in each section of the guide. Feel free to skip this step if you've already defined it in your session.
This page will step you through the reduction of the Antennae Band 7 SV data set for the Northern Mosaic using these 'for' loops. We will not be able to show every diagnostic plot but we give an example of each and the syntax to generate the rest. Also please be aware that even on a very fast machine this whole process can take a while, we are simply dealing with a lot of data.
A Priori Flagging
Even before we look in detail, we know that there are some data that we wish to exclude. We will start by flagging "shadowed" data where one antenna blocks the line of sight of another. We will also flag scans that were used to carry out pointing and atmospheric calibration, identified by their scan intent. Finally, we'll flag the autocorrelation data (the correlation of the signal from an antenna with itself) as we are only interested in cross-correlation data to make an interferometric image.
Start by defining our list of MS files:
# In CASA
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49"]
You may want to reset the flagging if you have tried this step before and are starting over though this is not necessary on your first time through. Do so using flagdata:
# In CASA
for asdm in basename_north:
print "Resetting flags for "+asdm
flagdata(vis=asdm+'.ms',mode='manualflag', unflag=T, flagbackup=F)
To carry out the flagging, we will assemble a set of flagging commands and then use the flagcmd task to execute them. These commands will be stored in a Python list of strings called myflags, each of which will be a specific flagging command. The operation of flagcmd is decribed by help flagcmd (with examples), and the syntax for the flagging command modes is given in help tflagdata.
First, we designate mode='shadow' to remove data from a given antenna when another antenna is in front of it and blocking part of its aperture.
# In CASA
myflags = ["mode='shadow'"]
The relevant calibration information has already been extracted from the pointing and atmospheric scans and we will not need them below. Next we set up a command string to flag the pointing scans using mode='manual' selecting on the scan intent corresponding to pointing:
# In CASA
myflags.append("mode='manual' intent='*POINTING*'")
Note that because the atmospheric calibration scans contain only TDM spectral windows, they will be removed automatically when we separate out the FDM data below.
Now we wish to flag the autocorrelation data:
# In CASA
myflags.append("mode='manual' autocorr=True")
Finally we execute the same flagging commands on each ms individually using flagcmd:
# In CASA
for asdm in basename_north:
print "Executing 'a priori' flags for "+asdm
flagcmd(vis=asdm+'.ms', inpmode='cmd', command=myflags, action='apply', flagbackup=True)
As an added step, we use flagmanager to save a copy of the flags we just made in the MS to reflect the current state, in case we ever need to get back to this stage.
# In CASA
for asdm in basename_north:
print "Backing up 'a priori' flags for "+asdm
flagmanager(vis = asdm+'.ms', mode = 'save', versionname = 'Apriori')
Now if ever there is a reason to roll back the flags to match the current version, called 'Apriori', the syntax would be:
# In CASA
for asdm in basename_north:
print "Restoring up 'a priori' flags for "+asdm
flagmanager(vis = asdm+'.ms', mode = 'restore', versionname = 'Apriori')
The flagmanager task will also allow you to view the saved flagging versions, including those created by running flagdata with flagbackup=T. For example try
# In CASA
flagmanager(vis='uid___A002_X2181fb_X49.ms', mode='list')
The output in the logger should list the Apriori flagging that we have applied. It will also indicate versions associated with any flagdata command where you did not set flagbackup=F. Other tasks, for example applycal, will also create flag version tables.
Create and Apply Tsys, WVR, and Antenna Position Calibration Tables
#In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
Tsys
The Tsys calibration gives a first-order correction for the atmospheric opacity as a function of time and frequency and associates weights with each visibility that persists through imaging.
Use gencal to create the Tsys calibration tables from the spectral windows with CALIBRATE_ATMOSPHERE intents in listobs (i.e. the TDM spectral windows with 128 channels), these are spectral windows 5 and 7.
#In CASA
#Removing existing Tsys information
os.system('rm -rf *tdm.tsys')
for asdm in basename_all:
print "Creating TDM Tsys Table for "+asdm
gencal(vis=asdm+'.ms',caltable=asdm+'.tdm.tsys',spw='5,7',caltype='tsys')
Later in the applycal stage this TDM Tsys table will be interpolated to the FDM (3840 channels per spw) science spectral windows 1 and 3.
Next we inspect the Tsys tables for the spectral window spw=5 with the task plotcal. We want to check that Tsys data have reasonable values and identify any unexpected features as a function of either time or frequency. To get an idea of sensible Tsys under average atmospheric observations consult the ALMA sensitivity calculator, accessible from http://www.almascience.org (via the "Documents & Tools" link).
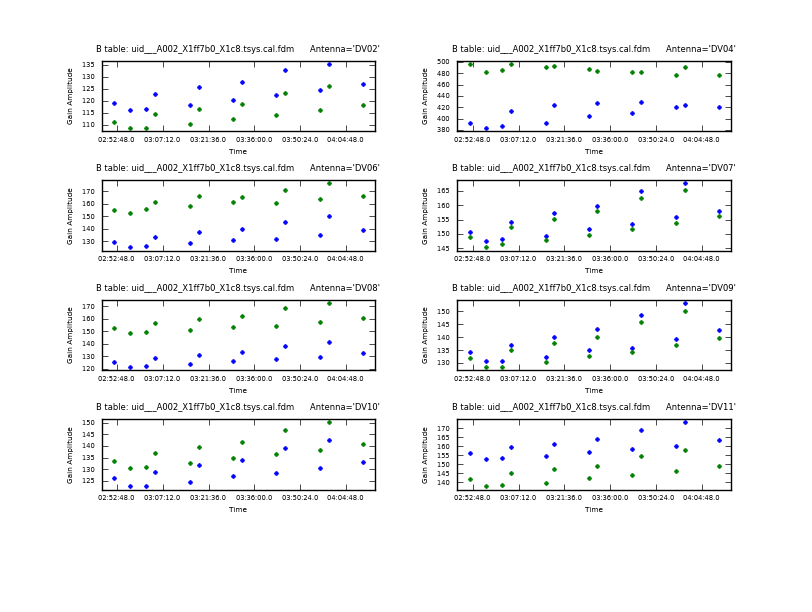
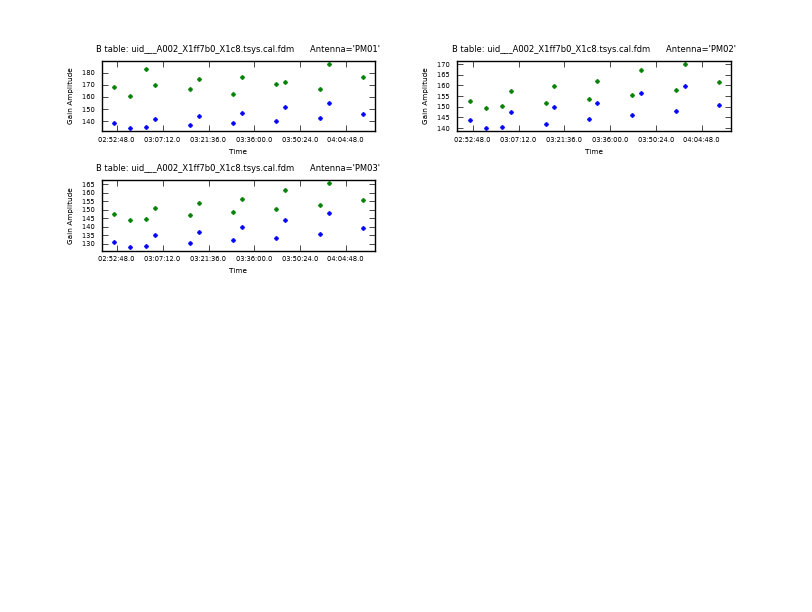
We start by plotting the Tsys for all the antennas and polarizations (XX and YY) as a function of time for each. Here and throughout we focus on spw 1, which contains CO(3-2): <figure id="uid___A002_X1ff7b0_X1c8.tsys_vs_time.page1.png">
</figure> <figure id="uid___A002_X1ff7b0_X1c8.tsys_vs_time.page2.png">
</figure>
#In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Plotting Tsys vs. time for "+asdm
plotcal(caltable=asdm+'.tdm.tsys',
xaxis="time",yaxis="tsys",
spw='5:50~50',plotsymbol=".", subplot=421,
antenna='0~7',
iteration='antenna', figfile=asdm+'.tsys_vs_time.page1.png',
fontsize=7.0)
#dummy_string = raw_input("First eight antennas for "+asdm+" . Hit <Enter> to continue.")
plotcal(caltable=asdm+'.tdm.tsys',
xaxis="time",yaxis="tsys",
antenna='8~15',
spw='5:50~50',plotsymbol=".", subplot=421,
iteration='antenna', figfile=asdm+'.tsys_vs_time.page2.png',
fontsize=7.0)
#dummy_string = raw_input("Remaining antennas for "+asdm+" . Hit <Enter> to continue.")
This sequence loops over all of our files and plots Tsys as a function of time for channel 50 in spectral window 5. In the call to plotcal:
- subplot=421 parameter sets up a 4 x 2 panel grid.
- iteration tells plotcal to make a separate plot for each antenna.
- spw '5:50~50' selects spw '5' channel 50-50. This allows us to cleanly separate time variations from frequency variations.
Because 8 panels (2 panels for each antenna - LSB and USB) is not enough to show all antennas on one page, there are two plotcal calls: one for the first 8 antennas (antenna=0~7), and then for the remaining antennas (antenna=8~15). The fontsize needs to be set to a small value or the text overlaps.
The 'raw_input' commands will wait for you to hit Enter before issuing the next plot command. In the example above these are commented out (the leading "#" means that CASA will ignore them). If you would like to interactively cycle through the plots, uncomment them by removing the "#". Otherwise, the figfile parameter directs the output to .png files for later inspection. The easiest way to look at the 20 plots produced here is to simply inspect the .png files using your favorite viewer.
The Tsys values in <xr id="uid___A002_X1ff7b0_X1c8.tsys_vs_time.page1.png"/> and <xr id="uid___A002_X1ff7b0_X1c8.tsys_vs_time.page2.png"/> look reliable, with typical values ~150 K except for some large values of Tsys at ~400 and 500 K for DV04. We will flag the data for that antenna later.
We will also want to look at Tsys as a function of frequency. This will use the analysisutils package mentioned at the beginning of this guide (called by the au. command)
#In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
os.system('rm -rf cal_plots/Tsys_plots/'+asdm+'.tdm.tsys*')
tsysfields=['3c279','Titan','Antennae']
caltable=asdm+'.tdm.tsys'
for field in tsysfields:
au.plotbandpass2(caltable=caltable,yaxis='amp',field=field,xaxis='freq',
showatm=True,overlay='time',
figfile='cal_plots/Tsys_plots/'+caltable+'.'+field,buildpdf=False,
interactive=False,chanrange='8~120',subplot=42)
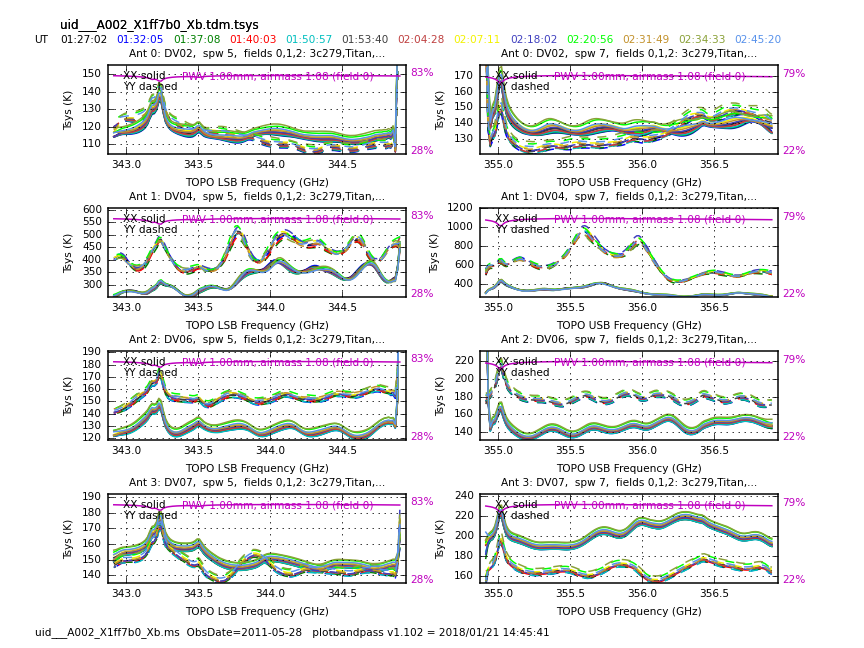
<figure id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.DV02.spw5.CASA3_4.png">
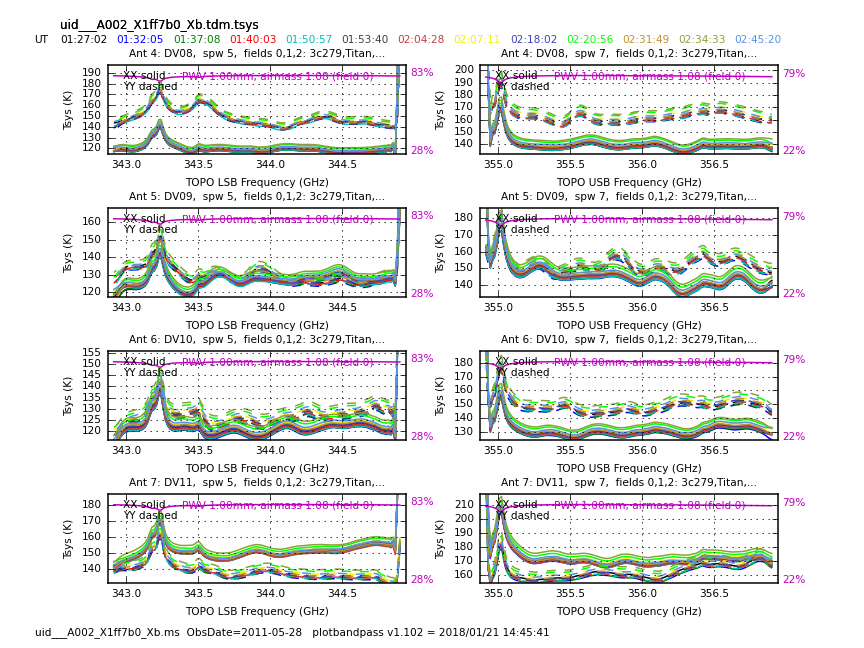
</figure> <figure id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.DV08.spw5.CASA3_4.png">
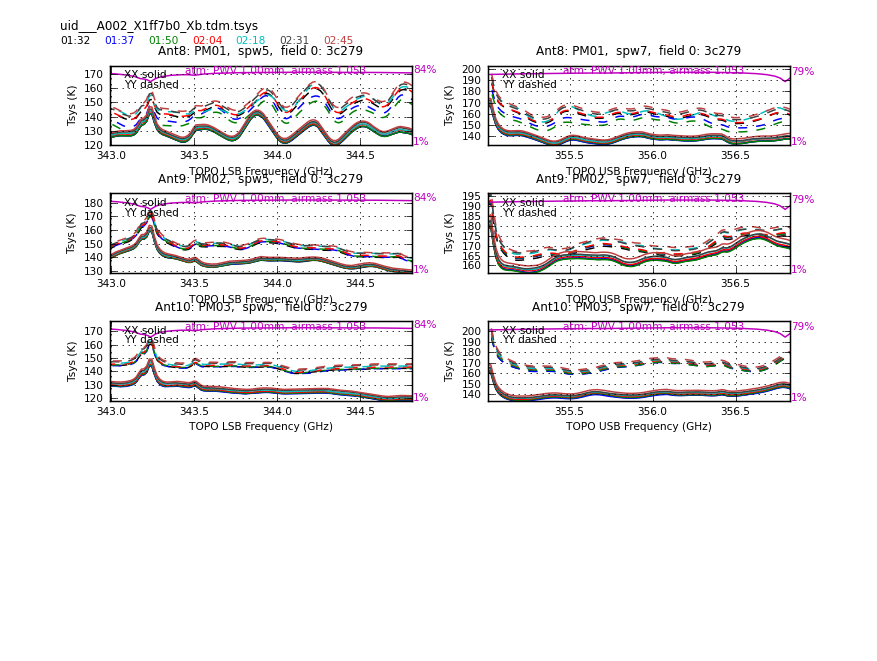
</figure> <figure id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.PM01.spw5.CASA3_4.png">
</figure>
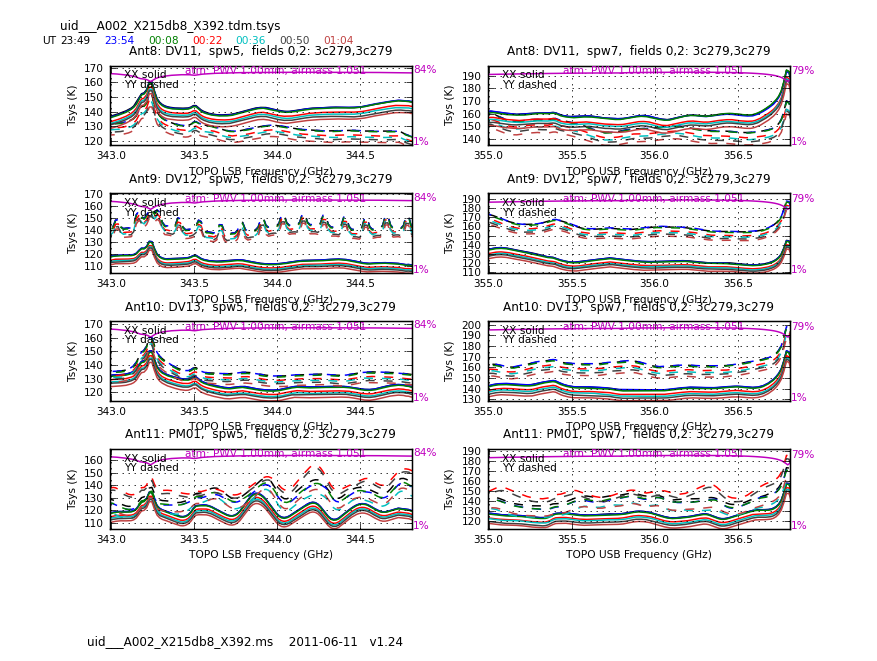
<figure id="Uid___A002_X215db8_X392.tdm.tsys.3c279.DV11.spw5.CASA3_4.png">
</figure>
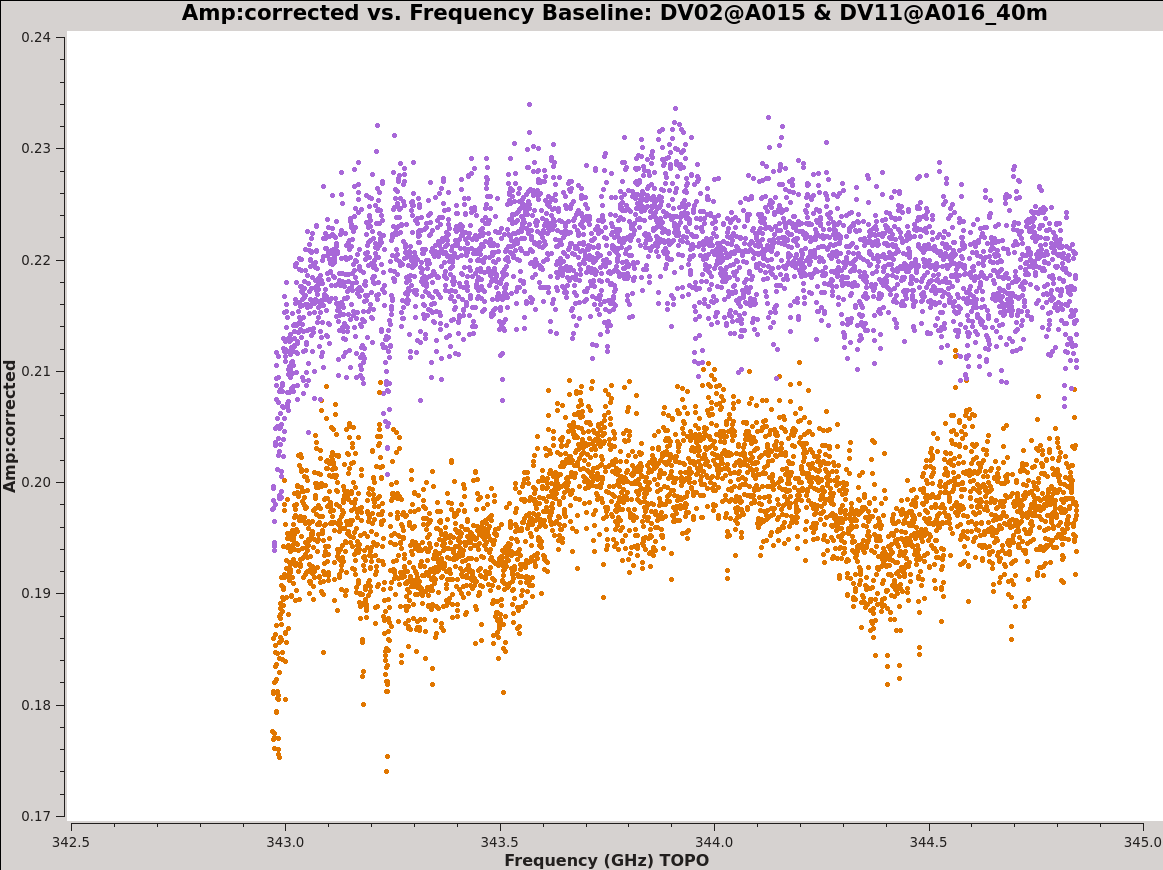
Now have a look at the Tsys vs. frequency plots or see <xr id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.DV02.spw5.CASA3_4.png"/>, <xr id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.DV08.spw5.CASA3_4.png"/>, and <xr id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.PM01.spw5.CASA3_4.png"/> for examples on the first data set. You can see the effect of a close pair of atmospheric ozone absorption lines at about 343.2 GHz that makes Tsys larger near that frequency in all antennas. Applying the Tsys calibration tables will minimize the contribution of these atmospheric lines. Again DV04 stands out with its very high Tsys. Although not present in the first data sets, Antenna DV12 exhibits periodic spikes in Tsys vs. frequency for one polarization (see <xr id="Uid___A002_X215db8_X392.tdm.tsys.3c279.DV11.spw5.CASA3_4.png"/> for an example from a later data set). It may or may not be possible to calibrate that behavior out. We will make a note to look carefully at DV12 further on in the calibration process.
WVR
The WVR calibration uses observations of the wings of the 183 GHz atmospheric water line to correct for phase variations as a function of time. As noted in the tsys section, this run of gencal will append to existing WVR produced tables. To avoid this, preexisting wvrgcal data is removed.
#In CASA
#Removing existing WVR information
os.system('rm -rf *.wvrgcal')
for asdm in basename_all:
wvrgcal(vis=asdm+'.ms',caltable=asdm+'.wvrgcal',toffset=-1)
Antenna Positions
The antenna position table reflects refinements in the measured positions of the antennas from those stored in the data. gencal will now be used put antenna position data into each observation. Again, gencal will merely append to existing antenna position data, ruining any subsequent results. We start by removing any existing antenna position refinements, followed by defining the antenna names, then their refinements (both as arrays), finally running gencal to create the information CASA can refer to for antenna positions.
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X1ff7b0_Xb.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:21]+'.antpos')
antenna='DV02,DV04,DV06,DV07,DV08,DV09,DV10,DV11,PM01,PM02,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00034, 0.00092, 0.00027, #PM02-A017 Before July 1
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X207fe4_X3a.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:22]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X207fe4_X3b9.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:23]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X2181fb_X49.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:22]+'.antpos')
antenna='DV01,DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,DV13,PM01,PM03'
parameter = [
-0.00004, 0.00026, 0.00019, #DV01-A137
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
0.00009, 0.00033, 0.00023, #DV13-A075
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X1ff7b0_X1c8.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:23]+'.antpos')
antenna='DV02,DV04,DV06,DV07,DV08,DV09,DV10,DV11,PM01,PM02,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00034, 0.00092, 0.00027, #PM02-A017 Before July 1
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X207fe4_X1f7.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:23]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X207fe4_X4d7.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:23]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV11,DV12,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X215db8_X18.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:22]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,DV13,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
0.00009, 0.00033, 0.00023, #DV13-A075
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X215db8_X1d5.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:23]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,DV13,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
0.00009, 0.00033, 0.00023, #DV13-A075
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
#In CASA
#The measurement set we are refining the antenna positions for
data='uid___A002_X215db8_X392.ms'
#Removing existing antenna refinements
os.system('rm -rf '+data[0:23]+'.antpos')
antenna='DV02,DV04,DV05,DV06,DV07,DV08,DV09,DV10,DV11,DV12,DV13,PM01,PM03'
parameter = [
0.00000, 0.00000, 0.00000, #DV02-A015
-0.00004, 0.00059, 0.00024, #DV04-J505
-0.00019,-0.00003, 0.00035, #DV05-A067
-0.00027, 0.00033, 0.00022, #DV06-T704
0.00006, 0.00057, 0.00032, #DV07-A004
-0.00027, 0.00025,-0.00003, #DV08-A072
-0.00037, 0.00025, 0.00003, #DV09-A008
-0.00056, 0.00028, 0.00026, #DV10-A009
-0.00028, 0.00024, 0.00011, #DV11-A016
-0.00018, 0.00010,-0.00014, #DV12-A011
0.00009, 0.00033, 0.00023, #DV13-A075
-0.00017, 0.00013, 0.00030, #PM01-T702
-0.00007, 0.00034, -0.00004] #PM03-J504
gencal(vis=data,caltable='%s.antpos'%(data.split('.')[0]),caltype='antpos',
antenna=antenna,parameter=parameter)
Applycal
We are now ready to apply the Tsys and the WVR calibration tables to the data with applycal, which reads the specified gain calibration tables, applies them to the (raw) data column, and writes the calibrated results into the corrected column. Again, we loop through all the datasets. It is important to only apply Tsys and WVR corrections obtained close in time to the data being corrected, so in addition to looping over data sets we define the list of unique source names and loop over these. Then by setting gainfield and field to the same value we ensure that Tsys and WVR calibrations are only applied to the source for which they are measured. Because the source has a different name in the Northern Mosaic and the Southern Mosaic, we will carry out two loops. We will only correct spw 1 and 3, our science windows, because we will drop the other data in a moment.
The applycal task now has much more flexibility for interpolating and applying calibrations derived in one spectral window to another, even if they do not share the same spectral shape (number of channels and channel width). This new functionality is used below to interpolate the TDM (128 channel) Tsys measurements to the FDM (3840 channel) spectral windows. This is controlled through the spectral window mapping parameter *spwmap*. Because this can be a bit confusing, we've written a "helper" function that will tell you what you should put for the Tsys calibration table part of spwmap. We only need to run it on one of the datasets because they are all the same in this regard
# In CASA
from recipes.almahelpers import tsysspwmap
tsysspwmap(vis='uid___A002_X207fe4_X3a.ms',tsystable='uid___A002_X207fe4_X3a.tdm.tsys')
This will print to your screen: [0, 5, 5, 7, 5, 5, 5, 7]. Recall that the TDM spectral windows are spw=5 and 7 and the FDM spectral windows are 1 and 3. This spwmap tells applycal to use the Tsys information in spectral window 5 for spectral window 1; and to use the information in spectral window 7 for spectral window 3. Beyond that, it is only important that there are placeholders for the other spectral windows, its also useful to note that you need not give placeholders for spectral window ids larger than the largest spectral window you need to use, in this case 7.
You will also need to put in placeholders for other tables that you give the gaintables. So if the Tsys table is 2nd in the list, spwmap=[[],[0,5,5,7,5,5,5,7],[]]
Now run the applycal commands.
# In CASA
# A new list of file names that contain only data from the Northern Mosaic
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9","uid___A002_X2181fb_X49"]
field_names_north = ['Titan','3c279','NGC*']
for asdm in basename_north:
print "Apply Tsys, WVR, and Antenna Position calibrations to "+asdm
for field in field_names_north:
applycal(vis=asdm+".ms", spw='1,3',
field=field, gainfield=['',field,field],
interp=['','linear,spline','nearest'],
gaintable=[asdm+'.antpos',asdm+'.tdm.tsys',asdm+'.wvrgcal'],
spwmap=[[],[0, 5, 5, 7, 5, 5, 5, 7],[]],
flagbackup=F)
# A new list of file names that contain only data from the Southern Mosaic
basename_south=["uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7","uid___A002_X207fe4_X4d7",
"uid___A002_X215db8_X18","uid___A002_X215db8_X1d5","uid___A002_X215db8_X392"]
field_names_south = ['Titan','3c279','Ant*']
for asdm in basename_south:
print "Apply Tsys, WVR, and Antenna Position calibrations to "+asdm
for field in field_names_south:
applycal(vis=asdm+".ms", spw='1,3',
field=field, gainfield=["",field,field],
interp=['','linear,spline','nearest'],
gaintable=[asdm+".antpos",asdm+'.tdm.tsys',asdm+'.wvrgcal'],
spwmap=[[],[0, 5, 5, 7, 5, 5, 5, 7],[]],
flagbackup=F)
where:
- field: the field to which we will apply the calibration,
- gainfield: the field from which we wish to take the calibration table
- interp = 'nearest' : apply the nearest solution from the calibration table rather than interpolating.
As you browse through the whole data set, you will probably note some problems along the same lines as the DV04 issue we saw above. We'll apply these as additional data flagging in just a moment.
Inspect Data
We are not quite done with the original ".ms" data sets yet. Before going further it will be useful to use plotms to show the effects of applying the calibration. In the process we'll take a quick look at each antenna and search for pathologies in the data.
For this basic inspection, we want to compare the phase and amplitude as a function of frequency and time in the DATA and CORRECTED columns of each measurement set. The CORRECTED column has had the Tsys and WVR calibrations applied and so we expect lower phase scatter and flatter amplitude response as a function of time and frequency. We are looking for antenna-based issues, so cycling through a set of baselines that includes each antenna once will be a good start. We'll focus these plots on the phase+bandpass calibrator, 3c279, and on baselines that include antenna DV11, which we will make our reference antenna in just a bit.
Each CASA Measurement Set has up to three "columns" of data: DATA, CORRECTED, and MODEL (though it is possible for a MS to hold only a DATA column if it has not been processed at all). A column entry exists for each data point (baseline, frequency, time). The DATA column holds the current raw data, though using split as we just did we can change the definition of "raw" data. The CORRECTED column holds the result of applying one or more calibration tables (e.g., via applycal) to the DATA column and so represents a corrected version of the DATA column. In CASA 3.4: the MODEL column is deprecated (though it is still possible to use them by explicitly setting usescratch=T). Instead, the model is stored in the header of the ms. This is a great step forward in terms of saving space and time to create scratch columns To get an intuitive feel for the columns you may wish to explore using plotms (which can plot different data columns, as seen below) or the browsetable task.
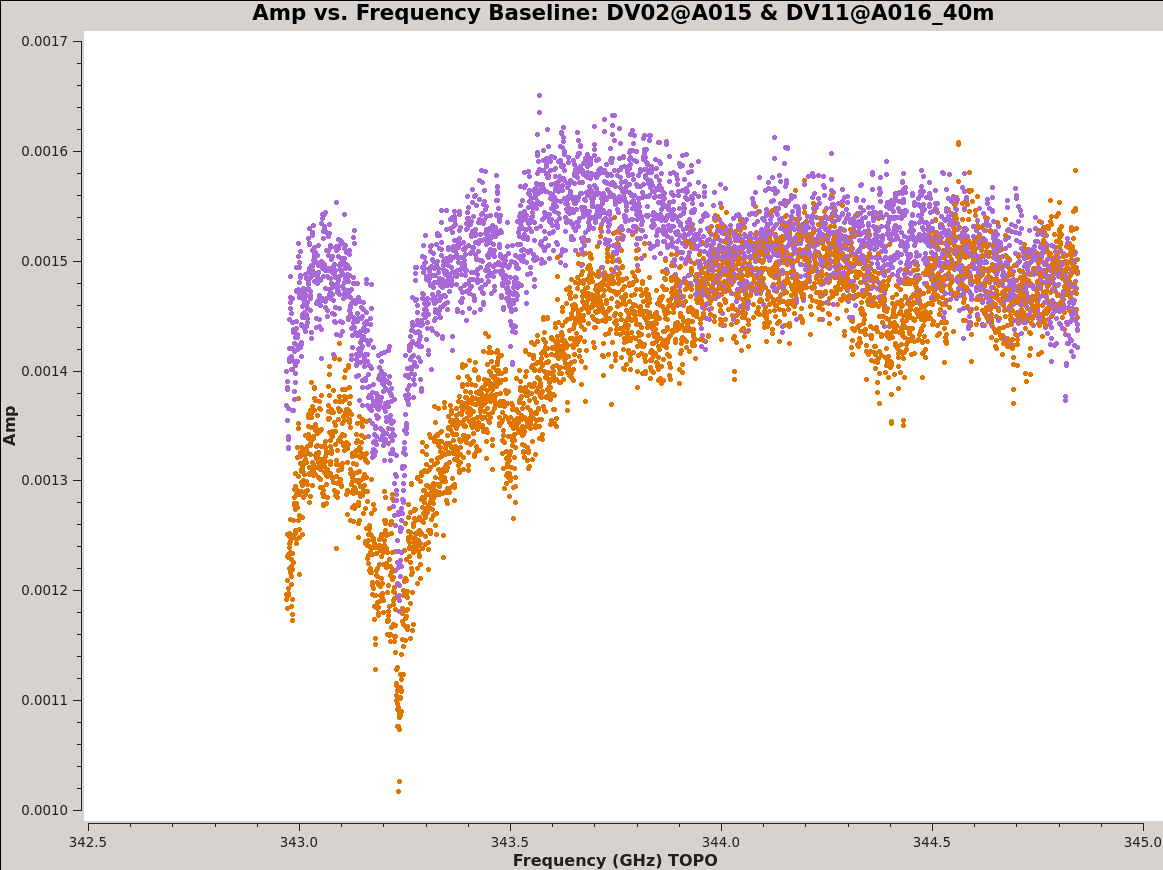
<figure id="plotms_amp_vs_freq_example-data.png">
</figure> <figure id="plotms_amp_vs_freq_example-corr.png">
</figure>
First, we plot amplitude as a function of frequency for 3c279. We start by plotting the DATA column, set color to indicate the two correlations (i.e., the XX and YY polarizations), and ask plotms to iterate over baseline. By setting antenna to 'DV11&*' we select only baselines that include DV11. We ask plotms to average all data over a very long timescale, avgtime = 1e8 seconds ~ 3 years or much longer than the time spanned by the whole data set. By setting avgscan = True we allow plotms to average across scan boundaries. The result is a plot of average amplitude per channel vs. frequency.
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
asdm=basename_all[0]
plotms(vis=asdm+'.ms',
field='3c279',
xaxis='frequency', yaxis='amp',
selectdata=T, spw='1',
avgtime='1e8',avgscan=T,
coloraxis='corr',
iteraxis='baseline',
antenna='DV11&*',
ydatacolumn='data')
Notice the green arrows along the bottom of the plotms window. We asked plotms to iterate over baseline. As you click the arrows, the plot will rotate from baseline to baseline, always with DV11 so that each antenna shows up once. To see the effect of the calibration, go to the "Axes" tab along the left of the plotms window and pull down the Data Column menu under the Y Axis. Set this from DATA to CORRECTED and you should see the effects of the calibration. You may need to ensure that the "Force Reload" box is checked before clicking "Plot" (both buttons lie at the bottom of the panel). For the most part things get better (flatter), but as we noted before DV04 is problematic.
<figure id="plotms_amp_vs_time_north.png">
</figure> <figure id="plotms_amp_vs_time_south.png">
</figure>
You can now make analogous calls to examine the phase vs. frequency, amplitude vs. time, and phase vs. time.
# In CASA
plotms(vis=asdm+'.ms',
field='3c279',
xaxis='frequency', yaxis='phase',
selectdata=T, spw='1',
avgtime='1e8',avgscan=T,
coloraxis='corr',
iteraxis='baseline',
antenna='DV11&*',
ydatacolumn='data')
plotms(vis=asdm+'.ms',
field='3c279',
xaxis='time', yaxis='amp',
selectdata=T, spw='1:1200~1300',
avgchannel='1000',avgscan=F,
coloraxis='corr',
iteraxis='baseline',
antenna='DV11&*',
ydatacolumn='data')
plotms(vis=asdm+'.ms',
field='3c279',
xaxis='time', yaxis='phase',
selectdata=T, spw='1:1200~1300',
avgchannel='1000',avgscan=F,
coloraxis='corr',
iteraxis='baseline',
antenna='DV11&*',
ydatacolumn='data')
Where:
- spw is set to cover only channels 1200~1300 for the time plots in order to isolate time dependent variations from frequency-dependent behavior. Those 101 channels represent only a small part of the total spw 1 bandpass.
- avgchannel set to a large number causes the plots of phase and amplitude vs. time to average data at all frequencies into a single point for each measurement.
- coloraxis corr sets the colors to correspond to the two polarizations of the data.
In each case, you will want to examine each baseline, alternating between the DATA and CORRECTED columns.
This is a lot of data inspection and that's only for one of 10 data sets! You can iterate across the data by hand, updating "asdm" to refer to each data set in order and cycling between baselines and DATA/CORRECTED. It is also possible to script CASA to show you the key plots in succession (see the next block down). However you approach the infrastructure, you are looking for:
- Improved scatter and lower variability in phase and amplitude vs. frequency and time. This indicates that the WVR and Tsys calibrations helped.
- Sudden jumps in phase or amplitude as a function of either time or frequency. These may indicate problems with the antenna during that observation.
- Large gradients, especially full wraps, in phase as a function of frequency. This may indicate a problem in the delays, the signal path length to the telescopes.
- Unusual magnitude, scatter, or patterns in any plot - though this may be better explored using plots that show all data together, which we'll make in a moment.
- Missing data. For example, if the phase calibrator drops out for a period of time we will not be able to calibrate and will need to flag the data.
As you look through, note individual potentially problematic antennas. If all antennas in a data set appear problematic it may be that your "reference" antenna, DV11 in the example above, is the source of the problem. In this case swap this reference antenna for another and see whether the problem is isolated to your original reference antenna.
A brief aside on structure: This section (Data Inspection) and the next (Apply Flags) are closely linked. We will present them as two separate steps. The idea here is that you look through your data, note problems, and then write commands to flag problematic data. This mimics one approach to writing data reduction scripts for CASA, where you will group all flagging together into one convenient place. Other CASA guides take a different approach, interleaving flagging and plotting. There is no "right" answer here. Find an approach to data inspection that works for you.
If you do wish to semi-automate the plot generation, the following sequence will cycle between data and corrected plots for each data set in turn. Type "stop" at any input call to break out.
Note: In loops like this involving plotms, hitting enter before the last dataset completes loading may cause plotms to stop refreshing the plot window at each step, or cause the window to disappear altogether. This will be fixed in an upcoming CASA release. Right now, you may need to exit CASA and restart to bring up the plotms window again, but you should be able to continue in the script where you left off.
# In CASA
user_input = ""
for asdm in basename_all:
# check if a stop has been requested
if user_input == "stop":
break
# Extract antenna list for this data set.
tb.open(asdm+'.ms/ANTENNA', nomodify=True)
ants = tb.getcol('NAME')
tb.close
# Define the reference antenna to make baselines with
ref_ant = 'DV11'
# Loop over antennas
for ant in ants:
# Check if the user wants to stop
if user_input == "stop":
break
# Skip correlation of reference antenna with itself (autocorrelations are flagged anyhow)
if ant == ref_ant:
continue
# Define the baseline with the reference antenna for current antenna
ant_str = ref_ant+'&'+ant
print "Showing baseline "+ant_str+" for data set "+asdm
print "Use this to inspect effect of applying wvrcal and Tsys calibrations."
# Loop over phase and amplitude as axes
for y_axis in ["amp", "phase"]:
# Make 'before' plot for frequency x-axis
print "... "+y_axis+" vs. frequency for DATA:"
plotms(vis=asdm+'.ms', spw='1', field='3c279',
antenna=ant_str, xaxis="frequency", yaxis=y_axis,
avgtime="1e8", avgscan=T, coloraxis="corr",
ydatacolumn="data")
user_input = raw_input("Hit <ENTER> to see CORRECTED data [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
# Make 'after' plot for frequency x-axis
print "... "+y_axis+" vs. frequency for CORRECTED:"
plotms(vis=asdm+'.ms', spw='1', field='3c279',
antenna=ant_str, xaxis="frequency", yaxis=y_axis,
avgtime="1e8", avgscan=T, coloraxis="corr",
ydatacolumn="corrected")
user_input = raw_input("Hit <ENTER> to proceed to next plot [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
# Make 'before' plot for time x-axis
print "... "+y_axis+" vs. time for DATA:"
plotms(vis=asdm+'.ms', spw='1:1200~1300', field='3c279',
antenna=ant_str, xaxis="time", yaxis=y_axis,
avgchannel="1000", coloraxis="corr",
ydatacolumn="data")
user_input = raw_input("Hit <ENTER> to see CORRECTED data [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
# Make 'after' plot for time y-axis
print "... "+y_axis+" vs. time for CORRECTED:"
plotms(vis=asdm+'.ms', spw='1:1200~1300', field='3c279',
antenna=ant_str, xaxis="time", yaxis=y_axis,
avgchannel="1000", coloraxis="corr",
ydatacolumn="corrected")
user_input = raw_input("Hit <ENTER> to proceed to next plot [type 'stop'+<Enter> to break out].")
if user_input == "stop":
break
<figure id="plotms_amp_vs_uvdist_north.png">
</figure>
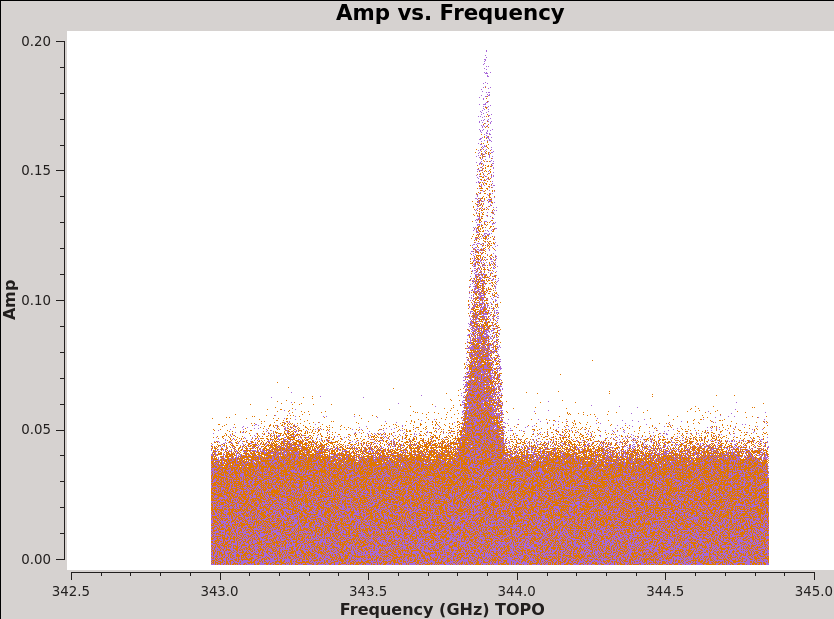
<figure id="plotms_amp_vs_freq_3c279.png">
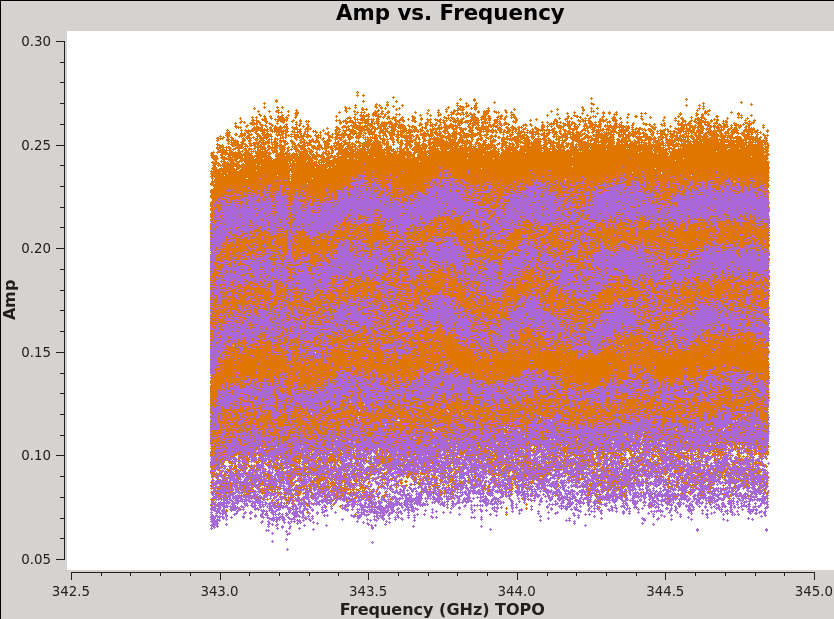
</figure> <figure id="plotms_amp_vs_freq_Titan.png">
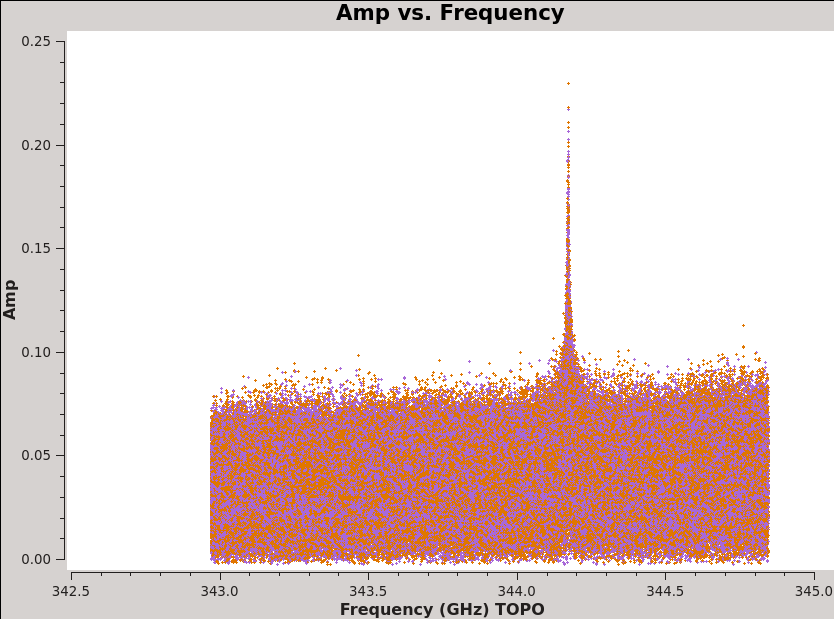
</figure> <figure id="plotms_amp_vs_freq_Antennae.png">
</figure>
A detailed explanation of the procedure is a bit outside the scope of this guide (for more on python see http://www.python.org/ and for more on the CASA toolkit see http://casa.nrao.edu/), but the basic process is to loop over each data set, baseline with the reference antenna (here DV11), and y-axis of interest (phase or amplitude) then plot the effect of the calibration vs. frequency and time for each combination. Running this to step through the data will give you about 200 "before and after" plots from which you could note a subset of problematic cases to be followed up by hand. Many other strategies to inspect the data are also viable.
With the Tsys and WVR calibrations applied successfully and the a priori flagging taken care of we will now split out the corrected data. We will keep only the corrected data, specified via datacolumn, and only spectral window 1, which contains the FDM (high spectral resolution) observations of the CO(3-2) line. Setting keepflags=F tells split not to carry over any fully flagged rows from the original data set to the new MS. We give the new MS files the extension ".wvrtsys.ms" to indicate that they have been corrected for WVR and Tsys effects. Because split will not overwrite existing files, we remove any previous versions of the new MS before beginning.
# In CASA
for asdm in basename_all:
os.system('rm -rf '+asdm+'.wvrtsys.ms')
print "Splitting out corrected data for "+asdm
split(vis=asdm+'.ms', outputvis=asdm+'.wvrtsys.ms',
datacolumn='corrected', spw='1', keepflags=F)
Be patient, split may take awhile. The WVR and Tsys-corrected data now sit in the DATA column of the new measurement sets, which have only one spectral window (now labeled spectral window 0 though it was spectral window 1 in the original data). You may wish to run listobs to illustrate the changes:
# In CASA
for asdm in basename_all:
os.system('rm '+asdm+'.wvrtsys.listobs.txt')
listobs(vis=asdm+'.wvrtsys.ms', listfile=asdm+'.wvrtsys.listobs.txt', verbose=True)
Note the new spectral window information:
2011-08-05 01:07:08 INFO listobs Spectral Windows: (1 unique spectral windows and 1 unique polarization setups) 2011-08-05 01:07:08 INFO listobs SpwID #Chans Frame Ch1(MHz) ChanWid(kHz)TotBW(kHz) Ref(MHz) Corrs 2011-08-05 01:07:08 INFO listobs 0 3840 TOPO 344845.586 488.28125 1875000 344908.33 XX YY
Next we will do a bit more inspection using plotms to look at whole data sets. This will help us identify missing data or look for egregious outliers.
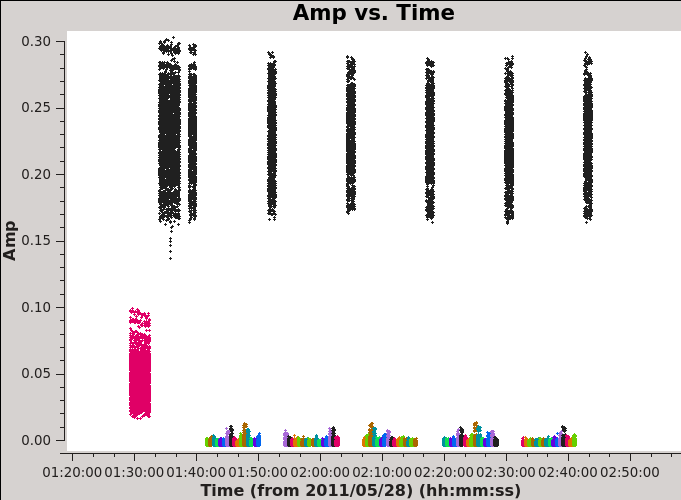
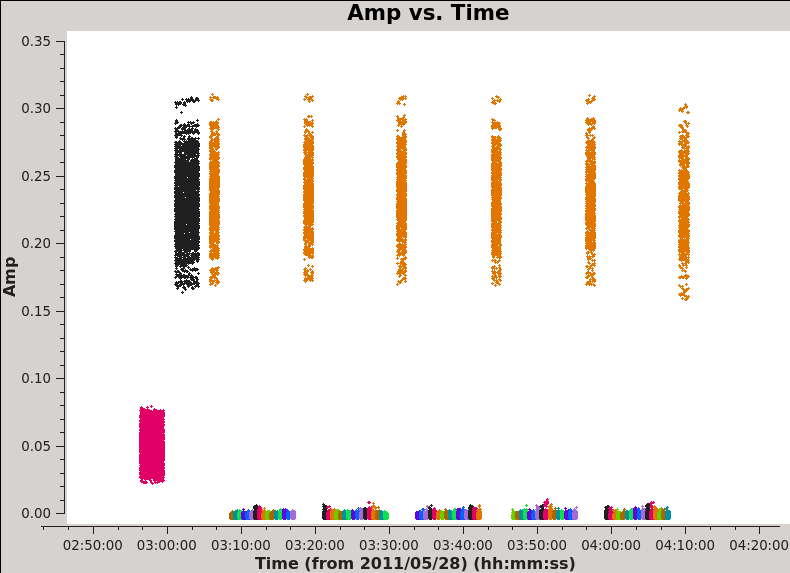
First we plot amplitude versus time (see <xr id="plotms_amp_vs_time_north.png"/> and <xr id="plotms_amp_vs_time_south.png"/>), averaging over all channels (by setting avgchannel to the very large value 10,000). We colorize by field so that scans on Titan are red, the bandpass and phase calibrator 3c279 is black (and orange in the Southern Mosaic where it has two field IDs), and the Antennae mosaic appears as a range of colors (one per pointing).
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
plotms(vis=asdm+'.wvrtsys.ms',
xaxis='time', yaxis='amp',
avgchannel='3840',coloraxis='field')
dummy_string = raw_input("Examining amplitude vs. time for "+asdm+" . Hit <Enter> to proceed.")
Here look for:
- Missing data. The source needs to be flanked by phase calibrator scans, if those are missing for any reason we need to flag the appropriate time range.
- Dramatic outliers. Does the source suddenly get very bright or the otherwise bright calibrator appear anomalously faint for a brief time? This likely indicates problematic data that should be identified and flagged. You can use the "select" (box with green plus along the bottom row in plotms) and "locate" (magnifying glass) buttons in plotms to isolate and identify problem data (it will print to the log).
- Smooth variation with time. A sudden jump may indicate a problem and often the safest approach is to flag data near a discontinuity.
Look through the amplitudes vs. time for each data set (remember that we've already examined the phases vs. time and amplitude vs. time for individual baselines above).
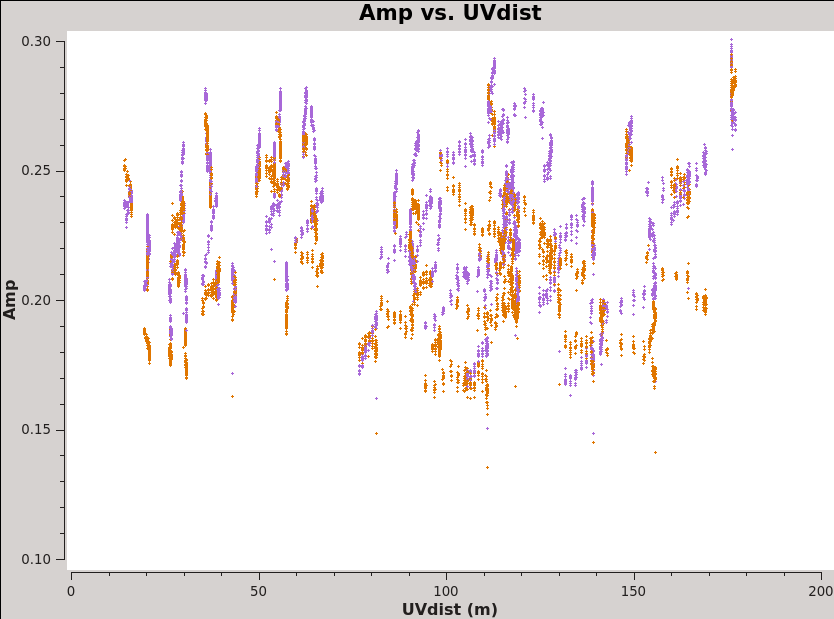
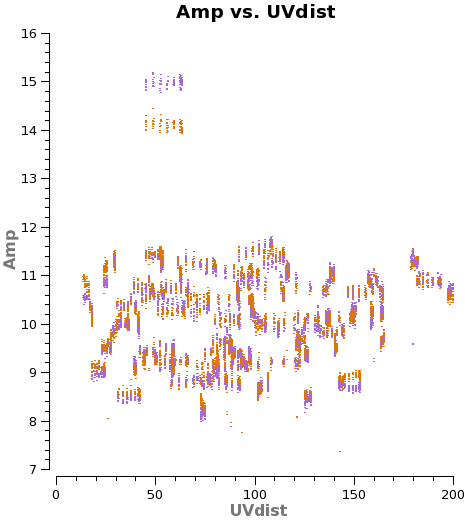
There are two other very useful "averaging" plots worth making. First, we plot amplitude as a function of u-v distance (projected antenna separation). Discontinuities and spikes in this plot are often from non-astrophysical sources. In the phase analog to the plot, the effects of atmospheric decorrelation can be assessed from increased scatter at longer u-v distances. While using the moon Titan as our flux calibrator, we may want to watch for flaring amplitudes at short u-v distances. These may indicate that Saturn is contaminating our beam. For a perfect, bright point source, we expect flat amplitudes as a function of u-v distance at the source amplitudes. <xr id="plotms_amp_vs_uvdist_north.png"/> shows an example of this plot, generated via:
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
plotms(vis=asdm+'.wvrtsys.ms',
field='3c279',
xaxis='uvdist', yaxis='amp',
avgchannel='3840',coloraxis='corr')
dummy_string = raw_input("Examining amplitude vs. UV Distance for 3c279 for "+asdm+" . Hit <Enter> to proceed.")
plotms(vis=asdm+'.wvrtsys.ms',
field='Titan',
xaxis='uvdist', yaxis='amp',
avgchannel='3840',coloraxis='corr')
dummy_string = raw_input("Examining amplitude vs. UV Distance for Titan for "+asdm+" . Hit <Enter> to proceed [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
It can also be useful to examine the average amplitude as a function of frequency for each target. This allows one to check for lingering atmospheric effects, unexpected line emission or absorption in the calibrators, or decreased sensitivity due to "roll-off" of the telescope sensitivity at the band edges. These plots are created by the sequence of commands below, where we have broken up the script to loop separately over the Northern and Southern Mosaics to reflect the different source names.
# In CASA
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9","uid___A002_X2181fb_X49"]
field_names = ["3c279","Titan","NGC*"]
for asdm in basename_north:
for field in field_names:
plotms(vis=asdm+'.wvrtsys.ms',
field=field,
xaxis='frequency', yaxis='amp',
avgtime='1e8',avgscan=T, coloraxis='corr')
dummy_string = raw_input("Examining amplitude vs. frequency for "+field+" in "+asdm+" . Hit <Enter> to proceed.")
basename_south=["uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7","uid___A002_X207fe4_X4d7",
"uid___A002_X215db8_X18","uid___A002_X215db8_X1d5","uid___A002_X215db8_X392"]
field_names = ["3c279","Titan","Ant*"]
for asdm in basename_south:
for field in field_names:
plotms(vis=asdm+'.wvrtsys.ms',
field=field,
xaxis='frequency', yaxis='amp',
avgtime='1e8',avgscan=T, coloraxis='corr')
dummy_string = raw_input("Examining amplitude vs. frequency for "+field+" in "+asdm+" . Hit <Enter> to proceed.")
For this command notice that we can see the CO(3-2) line in the Antennae even before calibration (see <xr id="plotms_amp_vs_freq_Antennae.png"/>) and that Titan also shows evidence of a strong line (also <xr id="plotms_amp_vs_freq_Titan.png"/>)! This will need to be flagged before we can use Titan to calibrate the flux scale of our data.
This suite of plots (along with the earlier inspection of the Tsys tables) gives us the tools we need to identify problematic data through the data sets. We use this to generate a set of inspection-driven flagdata commands for each data set. We apply these before the bandpass and gain calibration.
Apply Flagging
Based on this inspection and the other plots we have made, we now flag problematic portions of the data. We break up the flags by reason for flagging in order to illustrate the process. As you reduce your own data it may be more efficient to group flags by data set and make use of the flagcmd command. Except for the "post-calibration" flagging, the inspection plots that we just looked through have already revealed all of the problems that we flag. We structure the guide so that the flagging is grouped in one place.
As before, we may wish to reset our flags before beginning (particularly if one iterates this process) via:
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
flagdata(vis = asdm+'.wvrtsys.ms',mode='manualflag', unflag= T, flagbackup = F)
Remember that we dropped the flagged data when splitting out after the WVR and Tsys calibration, so this should not undo your "A Priori" flagging of shadowed data, autocorrelations, etc.. In any case this unflagging step is not necessary during the first iteration.
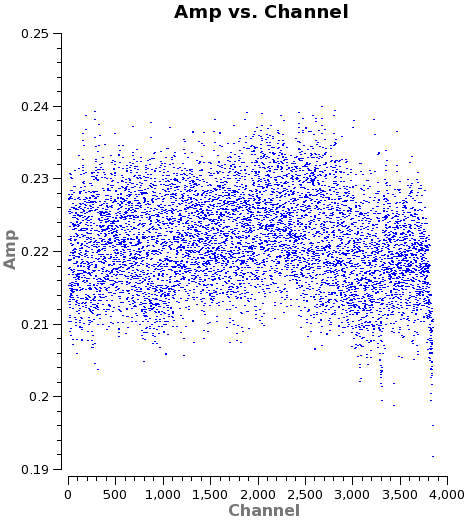
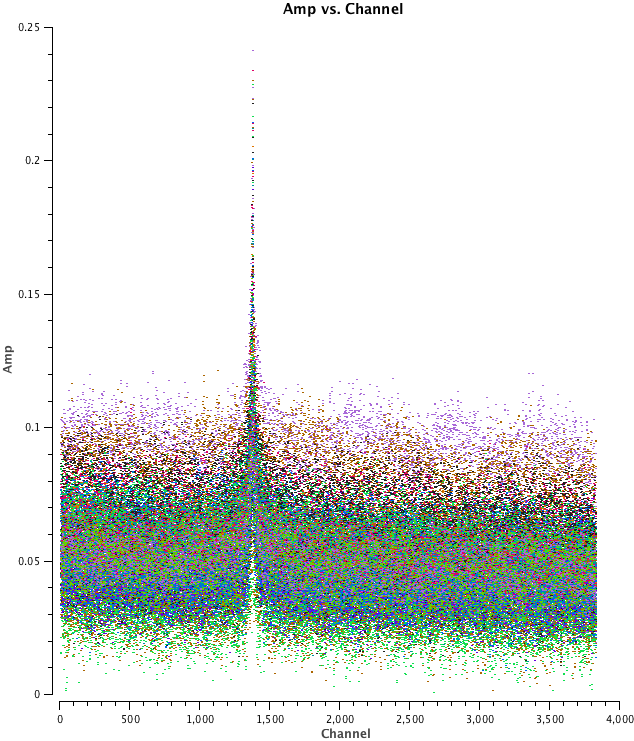
- Edge Channels
ALMA's sensitivity decreases near the edge of the baseband and it is often useful to check for a 'roll-off' in sensitivity near the edge of the band. This will appear as a downturn in amplitude as a function of channel near the edge of the window in the uncalibrated data, as a flaring due to increased noise at the spw edges in the calibrated data. It will also be visible in the amplitude part of the bandpass calibration table. Because our FDM spw does not cover the full baseband, we do not see a strong roll off in our data (see <xr id="amp_vs_channel_example.png"/>), where there is only a mild hint of a roll-off at the high end) but we do flag a (very) few channels at the high and low-frequency edge of the data set to be safe.
<figure id="amp_vs_channel_example.png">
</figure>
# In CASA
for asdm in basename_all:
flagdata(vis = asdm+'.wvrtsys.ms',spw = '0:0~7,0:3831~3839', flagbackup = F)
for asdm in basename_all:
flagdata(vis = asdm+'.wvrtsys.ms',spw = '0:3260~3320', flagbackup = F)
- Problematic Tsys measurements
Above we noted issues with the Tsys measurements for both DV04 and DV12. We flag the affected data. Each of these issues should be visible in the Tsys plots you made above (e.g., see <xr id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.DV02.spw5.CASA3_4.png"/> and <xr id="Uid___A002_X1ff7b0_Xb.tdm.tsys.3c279.PM01.spw5.CASA3_4.png"/>).
# In CASA
asdm="uid___A002_X1ff7b0_Xb"
flagdata(vis=asdm+'.wvrtsys.ms', antenna='DV04', flagbackup=F)
asdm="uid___A002_X207fe4_X3a"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X207fe4_X3b9"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X2181fb_X49"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X1ff7b0_X1c8"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV04',flagbackup=F)
asdm="uid___A002_X207fe4_X1f7"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X207fe4_X4d7"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X215db8_X18"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X215db8_X1d5"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
asdm="uid___A002_X215db8_X392"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV12',correlation='YY', flagbackup=F)
- Unreliable Short-Spacing Measurements on Titan
<figure id="amp_vs_uv_titan_obs.png">
</figure>
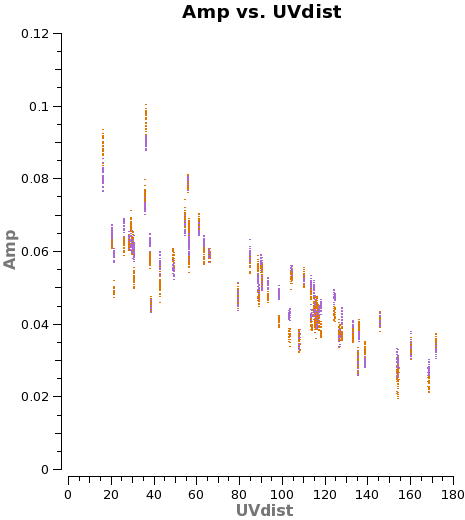
Saturn may contaminate the short u-v spacings from Titan. In any case these often show significant scatter (<xr id="amp_vs_uv_titan_obs.png"/>), so we flag them. There are still enough baselines to determine a good amplitude calibration for each antenna.
# In CASA
asdm="uid___A002_X1ff7b0_Xb"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~50', flagbackup = F)
asdm="uid___A002_X207fe4_X3a"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~40', flagbackup = F)
asdm="uid___A002_X207fe4_X3b9"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~50', flagbackup = F)
asdm="uid___A002_X2181fb_X49"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~30', flagbackup = F)
asdm="uid___A002_X1ff7b0_X1c8"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~50', flagbackup = F)
asdm="uid___A002_X207fe4_X1f7"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~30', flagbackup = F)
asdm="uid___A002_X207fe4_X4d7"
flagdata(vis=asdm+'.wvrtsys.ms', field='Titan', uvrange='0~30', flagbackup = F)
- Delay Issues
<figure id="phase_vs_freq_DV13.png">
</figure>
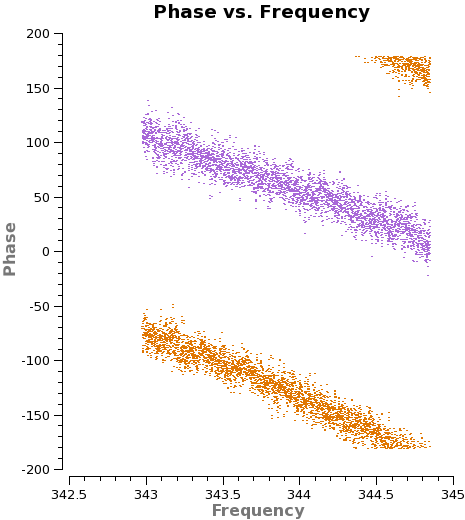
DV13 and a few other antennas show signatures of an imperfect delay calibration. This is most easily identified via strong "wrapping" of phase as a function of frequency (see <xr id="phase_vs_freq_DV13.png"/>). Such effects can be calibrated out with mild delay issues largely accounted for by the bandpass solution. The phase wrapping in DV13 seems weak enough that we will trust the calibrations to remove it. For a more extreme example see the CASA guide describing the calibration of NGC3256Band3.
- Missing Phase Calibrator Observations
<figure id="amp_vs_time_missing_scans.png">
</figure>
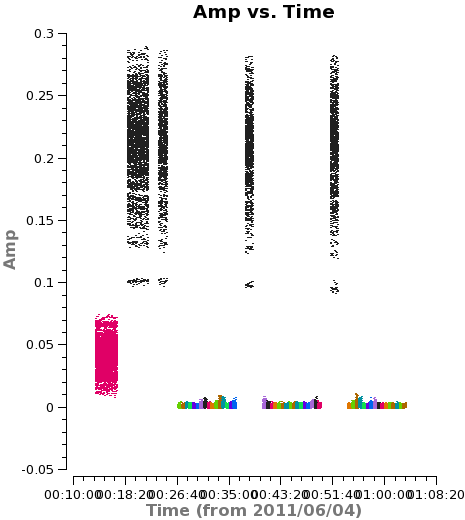
As a general rule, we want to be sure that observations of the phase calibrator (3c279) bracket each source observation. Two of the data sets do not include a final phase calibrator observation (see <xr id="amp_vs_time_missing_scans.png"/>) and for those two cases we flag the affected source observations.
# In CASA
asdm="uid___A002_X207fe4_X3b9"
flagdata(vis=asdm+'.wvrtsys.ms',timerange='00:53:47~01:08:00',flagbackup = F)
asdm="uid___A002_X215db8_X18"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV10',timerange='19:46:20~20:34:40',flagbackup=F)
- Unexpected Scatter or Discontinuity in the Data
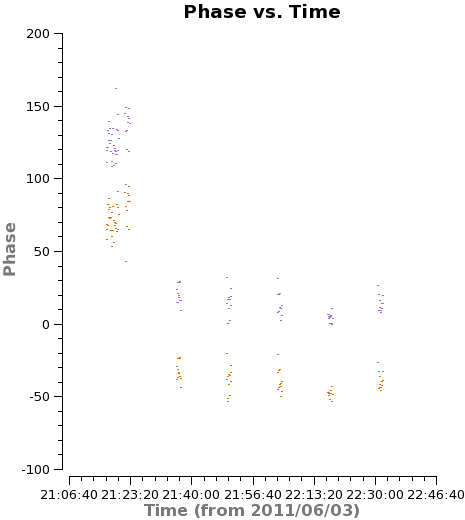
For several antennas we find sudden jumps in the phase of the phase calibrator as a function of time. These are visible in the plots of uncalibrated phase vs. time for single baselines above, and we show an example in <xr id="phase_vs_time_DV09.png"/>. It will not be possible to effectively interpolate the phase between measurements when we see these discontinuities. The safest approach is to flag the source data across these jumps. We do so here (though note that the last two flaggings are borderline cases).
<figure id="phase_vs_time_DV09.png">
</figure>
# In CASA
asdm="uid___A002_X207fe4_X3a"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV09', timerange='21:24:09~21:35:35', flagbackup = F)
asdm="uid___A002_X207fe4_X1f7"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV09',timerange='23:30:52~24:10:00',flagbackup=F)
flagdata(vis=asdm+'.wvrtsys.ms',antenna='PM01',timerange='23:16:50~24:10:00',flagbackup=F)
flagdata(vis=asdm+'.wvrtsys.ms',antenna='PM03',timerange='23:16:50~24:10:00',flagbackup=F)
- Outliers Visible After Calibration
<figure id="amp_vs_uvdist_outliers.png">
</figure>
Often issues with the data may become evident after calibration (i.e., after the next few steps that we apply). These data can appear as outliers in diagnostic plots for the calibrated data or even show up in the imaging stages. Once these are identified, best practice is to apply this new flagging then redo the calibration (if the issue is very minor, then re-calibrating may not be necessary).
# In CASA
asdm="uid___A002_X207fe4_X3a"
flagdata(vis=asdm+'.wvrtsys.ms',timerange='21:18:00~21:22:15', flagbackup=F)
asdm="uid___A002_X2181fb_X49"
flagdata(vis=asdm+'.wvrtsys.ms',antenna='DV01&DV04', flagbackup=F)
Now that we've applied our flagging, back up the flags as version "User" using flagmanager:
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
flagmanager(vis=asdm+'.wvrtsys.ms',mode='save',versionname ='User')
Applying this flagging will remove the most egregious pathologies from the data. We are now ready to calibrate the data.
Bandpass Calibration
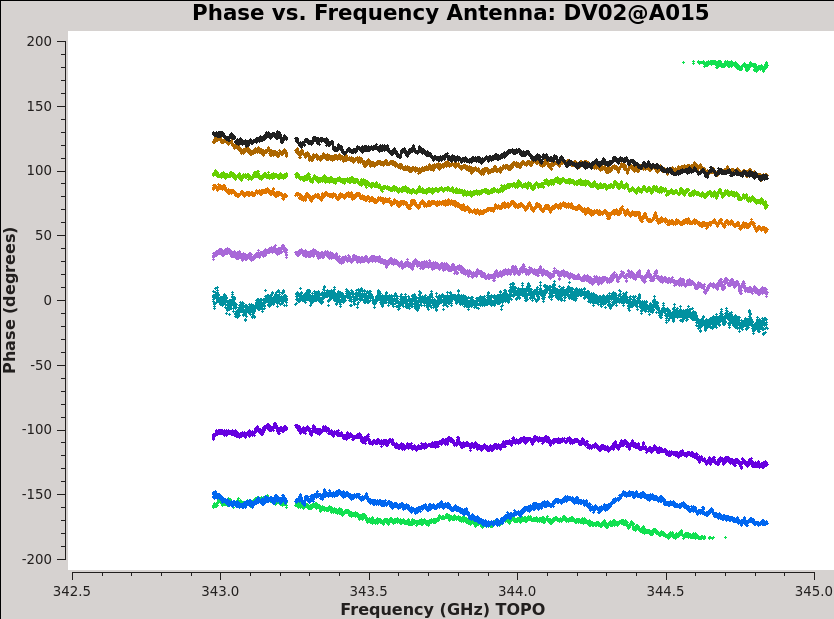
We begin by calibrating the phase and amplitude response of each antenna as a function of frequency, called "bandpass calibration." We have already seen that the data contain smooth but systematic variations in both phase and amplitude as a function of frequency. We can see this again in a more compact form by plotting phase as a function of frequency for all baselines associated with each antenna (<xr id="plotms_phase_vs_freq_example.png"/>).
<figure id="plotms_phase_vs_freq_example.png">
</figure>
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
plotms(vis= asdm+'.wvrtsys.ms',
xaxis='freq', yaxis='phase',
selectdata=True, field='3c279', correlation='XX',
avgtime='1e8', avgscan=T, antenna='*&*',
coloraxis='baseline', iteraxis='antenna')
dummy_string = raw_input("Plotting phase vs. frequency for "+asdm+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
Each plot shows phase as a function of frequency for all baselines with one antenna for 3c279. We plot only the 'XX' correlation, colorizing by baseline. With iteraxis set to antenna the green arrows at the bottom of plotms will cycle through antennas. By using avgscan and a large avgtime we average all scans and integrations.
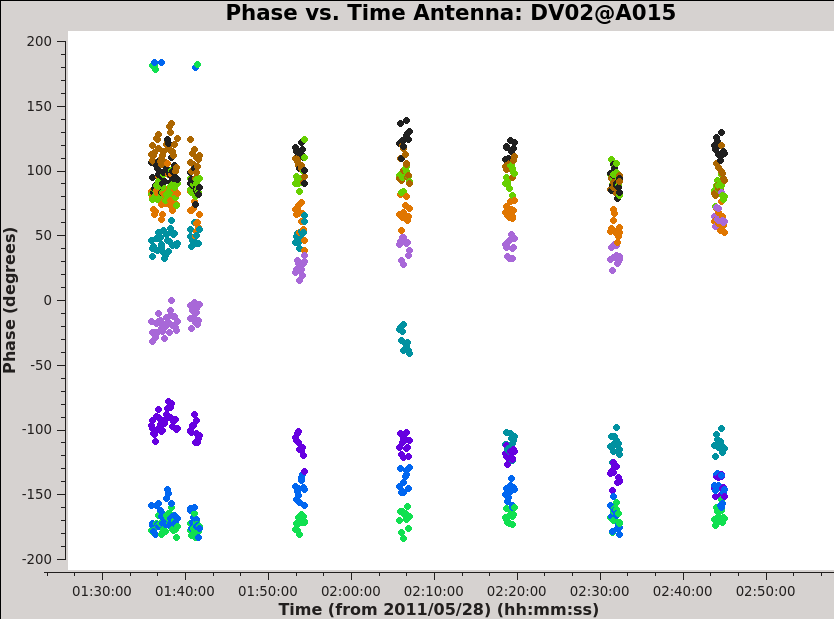
The phase (and amplitude) also varies as a function of time, as we saw before. Here are the similar plots for phase vs. time (see <xr id="Uid_A002_X1ff7b0_Xb-phasecont.png"/>).
<figure id="Uid_A002_X1ff7b0_Xb-phasecont.png">
</figure>
# In CASA
for asdm in basename_all:
plotms(vis= asdm+'.wvrtsys.ms',
xaxis='time', yaxis='phase',
selectdata=True, field='3c279',
spw='0:1200~1300', antenna='*&*',correlation='XX',
avgchannel='1000', avgscan=T,
coloraxis='baseline', iteraxis='antenna')
dummy_string = raw_input("Plotting phase vs. time for "+asdm+". Hit <Enter> for next data set or cycle through antennas [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
<xr id="Uid_A002_X1ff7b0_Xb-phasecont.png"/> shows that the phase varies with time. We need to take this temporal variation into account when we solve for the frequency variations. Therefore we carry out the bandpass calibration in two steps. First, we use gaincal to solve for the variation of phase as a function of time for 3c279 on very short timescales. We set gaincal to derive a separate phase solution for each antenna every integration by setting solint to 'int'. We solve, averaging together only a small fraction of the total bandpass (channels 1100-1300) to avoid the effects of the phase vs. frequency behavior. We will then apply this solution to remove time-dependent behavior when we solve for the frequency response of the antennas with bandpass.
An integration is the smallest time quantum of observation in the data, usually a few seconds. You can check the integration length for each scan in listobs.
#In CASA
for asdm in basename_all:
print "Running a short solution interval phase calibration for "+asdm
os.system('rm -rf '+asdm+'.bpphase.gcal')
gaincal(vis = asdm+'.wvrtsys.ms',
selectdata=T,field = '3c279',spw = '0:1100~1300',
caltable = asdm+'.bpphase.gcal',
solint = 'int',refant = 'DV11',calmode='p')
Now we use bandpass to solve for the frequency response of each antenna. To do this, we average all data in time by setting solint to 'inf' (that is, 'infinite'). We allow combination across scans and the different field IDs found for 3c279 by setting combine to "scan,field". We apply the phase vs. time calibration that we just derived on-the-fly using the parameter "gaintable".
for asdm in basename_all:
print "Running a bandpass calibration for "+asdm
os.system('rm -rf '+asdm+'.bandpass.bcal')
bandpass(vis = asdm+'.wvrtsys.ms',
field = '3c279',
gaintable = asdm+'.bpphase.gcal',
caltable = asdm+'.bandpass.bcal',
bandtype='B',
solint = 'inf',combine = 'scan,field', solnorm=T,refant = 'DV11',
minblperant=3,minsnr=2,fillgaps=62)
in these calls:
- caltable specifies the output calibration table
- gaintable specifies any calibration tables to be applied "on the fly" to the data before solving.
- solint sets the time interval for which solutions are derived for each antenna.
- refant set the reference antenna, here DV11, which is defined to have zero correction by construction.
- calmode 'p' sets gaincal to calibrate the phase only, other options are 'a'mplitude or 'ap' - amplitude and phase.
- minblperant sets the minimum baselines required per antenna for a successful solution.
- minsnr sets the minimum signal-to-noise ratio required for a successful solution.
- bandtype here 'B' tells bandpass to solve for the phase and frequency response of each antenna in each channel, averaging data in time across the solution interval.
- fillgaps tells bandpass to interpolate across channel gaps 1 channel wide.
- solnorm tells bandpass to normalize the bandpass amplitude and phase corrections to have magnitude unity. The absolute scale of the calibration will come from our later gaincal solutions.
Do not worry about the message "Insufficient unflagged antennas" when running the bandpass task. This indicates that bandpass is failing on the flagged edge channels, which is expected.
It is now a good idea to plot both sets of solutions to look for irregularities, especially:
- discontinuities in the phase vs. time solution
- rapid wrapping of phase in either phase vs. time or bandpass solution
- large roll-off in the amplitude response near the band edge in the bandpass solution
- large scatter in any solution.
We loop through and using analysis utils, again generating .png files of each calibration. As the bandpass plots take a while you may want to go have a cup of coffee and inspect them using your favorite image viewer. Otherwise, switch the interactive parameter to True to see them in real time instead.
<figure id="uid___A002_X1ff7b0_X1c8.bpphase.page1.png">
</figure> <figure id="Uid___A002_X1ff7b0_X1c8.bandpass.DV02.spw0.t1.CASA3.4.png">
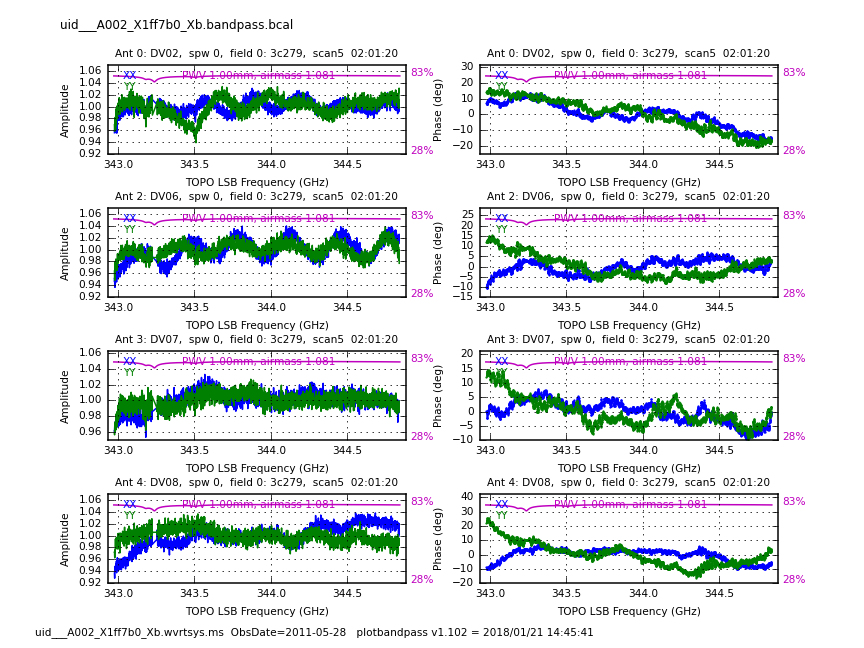
</figure> <figure id="uid___A002_X1ff7b0_Xb.bcal_phase.page1.png">
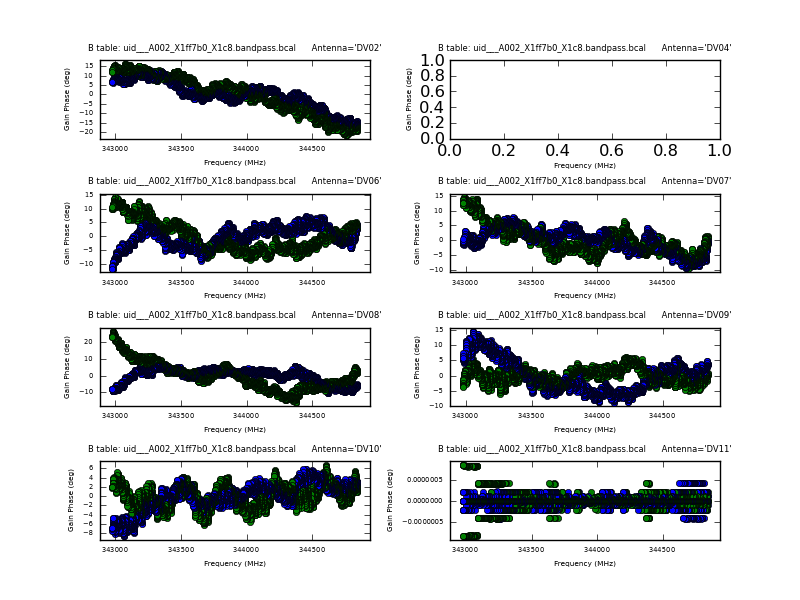
</figure>
#In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
os.system('rm -rf cal_plots/*bandpass*.png')
for asdm in basename_all:
au.plotbandpass2(caltable=asdm+'.bandpass.bcal',
field='',xaxis='freq',yaxis='both',
figfile='cal_plots/'+asdm+'.bandpass.png',
showatm=True,interactive=False,subplot=42)
Gain (Phase and Amplitude) Calibration
The bandpass calibration will account for the phase and amplitude response of our antennas as a function of frequency. We now solve for the absolute flux scale of the data by referencing to Titan and then calibrate the phase and amplitude behavior of the antennas as a function of time.
Before using Titan to set the flux, there is an important systematic to account for. When we looked at the integrated spectra of our targets above, remember that Titan showed a whopping spectral line, in fact the same CO(3-2) line that we wish to observe in the Antennae. We will set the flux of Titan (and thus all of our data) by referencing to a model in casa that does not account for this line. Therefore we need to flag the part of the Titan observations contaminated by the line before we calibrate (you can see the affected channel range in <xr id="Antennae-TitanLine.png"/>). We run the following additional flagging step:
<figure id="Antennae-TitanLine.png">
</figure>
# in CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Flagging CO(3-2) in Titan for "+asdm
flagdata(vis=asdm+'.wvrtsys.ms',flagbackup=F,
field=['Titan'],
spw=['0:1100~1700'])
for asdm in basename_all:
flagmanager(vis =asdm+'.wvrtsys.ms',mode = 'save',versionname = 'Calibration')
Next, we will use the setjy task to read the predicted complex visibilities for Titan into the MODEL column of each data set.
# in CASA
for asdm in basename_all:
print "Reading model for Titan into "+asdm
setjy(vis = asdm+'.wvrtsys.ms',field = 'Titan',
usescratch=False,scalebychan=True,
standard = 'Butler-JPL-Horizons 2010')
setjy will output the flux of Titan to the CASA logger and it is worth recording this information. The flux of Titan at these frequencies is about 2.9 Jy. For example, for dataset uid___A002_X1ff7b0_Xb.wvrtsys.ms:
2012-05-10 15:46:24 INFO setjy Titan (fld ind 1) spw 0 [I=2.8614, Q=0, U=0, V=0] Jy, (JPL-Butler Solar System Object)
Now we are going to take a look at the titan model for one dataset
# in CASA
plotms(vis ='uid___A002_X1ff7b0_X1c8.wvrtsys.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='model', field='Titan',avgtime='1e8',
avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr')
plotms(vis ='uid___A002_X1ff7b0_X1c8.wvrtsys.ms', xaxis='uvdist', yaxis='phase',
ydatacolumn='model', field='Titan',avgtime='1e8',
avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr')
Next we'll run a short-solution interval gaincal to solve for phase variation on short timescales during observations of our two calibrators, with solint set to "int". By applying this on-the-fly, we can remove any decorrelation in the data due to phase scatter when we solve for the amplitude calibration. However, there is no benefit to using this short-timescale solution to calibrate the source because we only have information on the gain during calibrator visits (though see the Imaging portion of this guide). Instead we will solve for the gains to apply to the source using a longer solint in just a moment.
# in CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Carrying out short timescale phase solution for "+asdm
os.system('rm -rf '+asdm+'.intphase.gcal')
gaincal(vis=asdm+'.wvrtsys.ms',
gaintable=asdm+'.bandpass.bcal',
caltable=asdm+'.intphase.gcal',
calmode='p',
field='Titan,3c279',
spw='0:40~3800',
refant='DV11', solint='int',minsnr=2.0, minblperant=4)
Now we derive the longer timescale phase calibration table using solint set to "inf", but not allowing scan combination. This calibration has higher signal to noise due to combining more data, and for the purposes of correcting the source, it is just as precise as the short timescale solution.
# in CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Carrying out longer timescale phase solution for "+asdm
os.system('rm -rf '+asdm+'.scanphase.gcal')
gaincal(vis=asdm+'.wvrtsys.ms',
gaintable=asdm+'.bandpass.bcal',
caltable=asdm+'.scanphase.gcal',
calmode='p',
field='Titan,3c279',
spw='0:40~3800',
refant='DV11', solint='inf',minsnr=2.0,minblperant=4)
Now we apply the short-timescale phase solution and carry out a scan length (solint set to "inf") calibration of the data using calmode of 'ap'.
# in CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Solving for longer (scan) interval amplitude solution for "+asdm
os.system('rm -rf '+asdm+'.amp.cal')
gaincal(vis = asdm+'.wvrtsys.ms',
gaintable =[asdm+'.bandpass.bcal',asdm+'.intphase.gcal'],
caltable = asdm+'.amp.cal',
calmode='ap',
field = 'Titan,3c279',
spw='0:40~3800',
refant = 'DV11',solint = 'inf', minblperant=4)
This "amp.cal" solution gives us the amplitude variations as a function of time, but they are not yet pinned to a realistic scale except in the case of Titan, where we have solved using the model input by setjy. We will set the flux of our secondary calibrator 3c279 with reference to Titan using fluxscale.
# in CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Scaling amplitude calibration to match Titan for "+asdm
os.system('rm -rf '+asdm+'.flux.cal')
fluxscale(vis = asdm+'.wvrtsys.ms',
caltable = asdm+'.amp.cal',
fluxtable = asdm+'.flux.cal',
reference = 'Titan',
transfer = '3c279')
This new correctly-scaled flux table ".flux.cal" replaces the previous ".amp.cal" table as the correct amplitude calibration table to apply to the data, i.e., the ".flux.cal" contains both the time variability of the amplitude solved for in ".amp.cal" and the correct flux scaling set with fluxscale.
Fluxscale will output the derived flux for 3c279 to the CASA logger. This information is worth noting. We find that the flux of 3c279 is 10.45 Jy, by averaging the fluxes obtained from the ten available datasets. This value agrees within 10% with the most recent 0.850 millimeter measurements from the SMA calibrator list [1] : (01 Jul 2011, SMA 9.75 ± 0.49).
Now we plot the final phase and amplitude calibration tables for each data set. A well-behaved calibration table will show smooth variations as a function of time. Sudden jumps or wild variations among the antenna amplitude gains should prompt further investigation and possibly additional flagging.
# in CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Plotting solutions for "+asdm
plotcal(caltable = asdm+'.scanphase.gcal',
xaxis = 'time', yaxis = 'phase',
iteration = 'antenna', plotrange=[0,0,-180,180],
showgui=False, subplot=421, figfile=asdm+'.scanphase.page1.png',
antenna='0~7', fontsize=6.0)
# dummy_string = raw_input("Hit <Enter> to see next plot.")
plotcal(caltable = asdm+'.scanphase.gcal',
xaxis = 'time', yaxis = 'phase',
iteration = 'antenna', plotrange=[0,0,-180,180],
showgui=False, subplot=421, figfile=asdm+'.scanphase.page2.png',
antenna='8~15', fontsize=6.0)
# dummy_string = raw_input("Hit <Enter> to see next plot.")
plotcal(caltable = asdm+'.flux.cal',
xaxis = 'time',yaxis = 'amp',
plotrange = [0,0,0,0],
antenna='0~7', iteration='antenna',
showgui=False, subplot=421, figfile=asdm+'.flux.page1.png',
fontsize=6.0)
# dummy_string = raw_input("Hit <Enter> to see next plot.")
plotcal(caltable = asdm+'.flux.cal',
xaxis = 'time',yaxis = 'amp',
plotrange = [0,0,0,0],
antenna='8~15', iteration='antenna', fontsize=6.0,
showgui=False, subplot=421, figfile=asdm+'.flux.page2.png')
# dummy_string = raw_input("Hit <Enter> to see next plot.")
Apply the Calibrations and Inspect
Now we will use applycal to apply the bandpass, phase, and amplitude calibration tables that we generated in the previous sections to the data. We apply the solutions separately to the bandpass and secondary ("phase") calibrator 3c279, the flux calibrator Titan, and the target source. In most data sets, the bandpass and secondary calibrator will not be the same and this step would include one additional applycal.
#In CASA
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9","uid___A002_X2181fb_X49"]
for asdm in basename_north:
print "Applying calibrations for "+asdm
applycal(vis=asdm+'.wvrtsys.ms',field='3c279',
gaintable=[asdm+'.bandpass.bcal',asdm+'.intphase.gcal',asdm+'.flux.cal'],
interp=['nearest','nearest','nearest'],
gainfield=['3c279','3c279','3c279'],flagbackup=F,calwt=F)
applycal(vis=asdm+'.wvrtsys.ms',field='Titan',
gaintable=[asdm+'.bandpass.bcal',asdm+'.intphase.gcal',asdm+'.flux.cal'],
interp=['nearest','nearest','nearest'],
gainfield=['3c279','Titan','Titan'],flagbackup=F,calwt=F)
applycal(vis=asdm+'.wvrtsys.ms',field='NGC*',
interp=['nearest','linear','linear'],
gaintable=[asdm+'.bandpass.bcal',asdm+'.scanphase.gcal',asdm+'.flux.cal'],
gainfield=['3c279','3c279','3c279'],flagbackup=F,calwt=F)
basename_south=["uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7","uid___A002_X207fe4_X4d7",
"uid___A002_X215db8_X18","uid___A002_X215db8_X1d5","uid___A002_X215db8_X392"]
for asdm in basename_south:
print "Applying calibrations for "+asdm
applycal(vis=asdm+'.wvrtsys.ms',field='3c279',
gaintable=[asdm+'.bandpass.bcal',asdm+'.intphase.gcal',asdm+'.flux.cal'],
interp=['nearest','nearest','nearest'],
gainfield=['3c279','3c279','3c279'],flagbackup=F,calwt=F)
applycal(vis=asdm+'.wvrtsys.ms',field='Titan',
gaintable=[asdm+'.bandpass.bcal',asdm+'.intphase.gcal',asdm+'.flux.cal'],
interp=['nearest','nearest','nearest'],
gainfield=['3c279','Titan','Titan'],flagbackup=F,calwt=F)
applycal(vis=asdm+'.wvrtsys.ms',field='Ant*',
interp=['nearest','linear','linear'],
gaintable=[asdm+'.bandpass.bcal',asdm+'.scanphase.gcal',asdm+'.flux.cal'],
gainfield=['3c279','3c279','3c279'],flagbackup=F,calwt=F)
Here:
- vis and field specify the measurement set and field to calibrate.
- gaintable specifies the tables to apply to the data. There may be several tables applied all at once, here we apply the bandpass, phase, and flux calibrations.
- interp specifies the type of interpolation used to match calibration tables to the data. If there are several gaintables then interp may be either a single value (e.g., "linear" or "nearest") or a list of values with size equal to the list of gaintables. In the case where interp is a list, applycal will match each element to the corresponding gaintable (e.g., first gaintable with first interp parameter).
- gainfield specifies field from which to draw the calibration. As with interp, this can be a list of size matched to gaintable, so that the first element of gainfield will be the field used for the first table. For example, in the above commands the bandpass calibration (the first table) will always be applied from '3c279' (the first gainfield).
- calwt indicates that the weights, which were derived from the Tsys measurements, should also be calibrated. Note that calwt is True by default.
Once calibrations are applied, it is important to go back and inspect the calibrated data. New problematic antennas or baselines may be visible after calibration. Repeat the steps above, focusing on the CORRECTED data column. Bear in mind that for any point source calibrators we now expect to find phase scattering around zero and to find flat amplitudes as a function of u-v distance. Look for outliers and other signatures of problematic data. As a general rule, you will want to incorporate these data into your overall flagging script then rerun the whole calibration process, so that reduction is iterative. If the data only represent a minor problem, however, it may not be terribly harmful to flag them after the fact so that they do not interfere with imaging but trust that the calibrations are mostly unaffected.
As an example of this inspection, we cycle through the corrected amplitudes and phases of 3c279 and Titan as a function u-v distance, to check that the phases are close to zero and the amplitudes are constant.
<figure id="xb-checkcal-pha-cha.png">
</figure> <figure id="xb-checkcal-amp-time.png">
</figure>
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Examining calibrated data for "+asdm
plotms(vis = asdm+'.wvrtsys.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', field='3c279',
averagedata=True, avgchannel='3840', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='time', yaxis='amp',
ydatacolumn='corrected', field='3c279',
averagedata=True, avgchannel='3840', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', field='3c279',avgtime='1e8',
avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='time', yaxis='phase',
ydatacolumn='corrected', field='3c279',
avgchannel='3840', avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
for asdm in basename_all:
print "Examining calibrated data for "+asdm
plotms(vis = asdm+'.wvrtsys.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', field='Titan',
averagedata=True, avgchannel='3840', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='uvdist', yaxis='phase',
ydatacolumn='corrected', field='Titan',
averagedata=True, avgchannel='3840', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', field='Titan',avgtime='1e8',
avgchannel='', avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot [type 'stop'+<Enter> to break out].")
# check if a stop has been requested
if dummy_string == "stop":
break
In <xr id="xb-checkcal-pha-cha.png"/> and <xr id="xb-checkcal-amp-time.png"/> we plot phase vs. channel and amp vs. time for 3c279 for the uid___A002_X1ff7b0_Xb dataset.
Finally we can use plotms to examine the corrected amplitude and phase of Antennae galaxies as a function of time and uv-distance (plots not shown):
# In CASA
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9","uid___A002_X2181fb_X49"]
for asdm in basename_north:
print "Examining calibrated data for "+asdm
plotms(vis = asdm+'.wvrtsys.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', field='NGC*',
averagedata=True, avgchannel='3840', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', field='NGC*',
avgtime='1e8', avgscan=T, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
basename_south=["uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7","uid___A002_X207fe4_X4d7",
"uid___A002_X215db8_X18","uid___A002_X215db8_X1d5","uid___A002_X215db8_X392"]
for asdm in basename_south:
print "Examining calibrated data for "+asdm
plotms(vis = asdm+'.wvrtsys.ms', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', field='Ant*',
averagedata=True, avgchannel='3840', avgtime='',
avgscan=F, avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
plotms(vis = asdm+'.wvrtsys.ms', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', field='Ant*',
avgtime='1e8', avgscan=T,avgbaseline=F, coloraxis='corr')
dummy_string = raw_input("Hit <Enter> for next plot.")
Split and Concatenate Data for Northern and Southern Mosaics
The individual data sets are now calibrated. We can safely split out the calibrated data for our science target and drop the calibrators. As we do so, we will smooth the data in frequency, averaging together groups of 23 channels by setting width=23 in split. The new data will have a channel width corresponding to about ~10 km/s, very similar to the SMA data being verified. The factor of >20 drop in data volume will also make the imaging steps much more tractable.
#In CASA
basename_north=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9","uid___A002_X2181fb_X49"]
basename_south=["uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7","uid___A002_X207fe4_X4d7",
"uid___A002_X215db8_X18","uid___A002_X215db8_X1d5","uid___A002_X215db8_X392"]
for asdm in basename_north:
os.system('rm -rf '+asdm+'.cal.ms')
split(vis = asdm+'.wvrtsys.ms',outputvis = asdm+'.cal.ms',
field = 'NGC*',spw='0',width=23, keepflags=False)
os.system('rm -rf '+asdm+'.cal.listobs.txt')
listobs(asdm+'.cal.ms',listfile=asdm+'.cal.listobs.txt')
for asdm in basename_south:
os.system('rm -rf '+asdm+'.cal.ms')
split(vis = asdm+'.wvrtsys.ms',outputvis = asdm+'.cal.ms',
field = 'Ant*',spw='0',width=23, keepflags=False)
os.system('rm -rf '+asdm+'.cal.listobs.txt')
listobs(asdm+'.cal.ms',listfile=asdm+'.cal.listobs.txt')
For convenience we concatenate all data for the Northern Mosaic into a single big MS and place all data for the Southern Mosaic into another file. To do this, we construct a list that holds the names of all the Southern Mosaic MS files and another that holds the name of all the Northern Mosaic MS files then feed these into the concat task.
Before we do this, we'll take a brief aside to remove Pointing information from the calibrated data. Normally, the pointing information would apply important corrections to be used while imaging the data. However, some aspects of this are still under development and the pointing information in these data interacts poorly with CASA. We will remove the pointing information using the CASA toolkit Table tool (tb). This kind of operation is dangerous and should not be a regular feature of your CASA reduction.
# In CASA
basename_all=["uid___A002_X1ff7b0_Xb","uid___A002_X207fe4_X3a","uid___A002_X207fe4_X3b9",
"uid___A002_X2181fb_X49","uid___A002_X1ff7b0_X1c8","uid___A002_X207fe4_X1f7",
"uid___A002_X207fe4_X4d7","uid___A002_X215db8_X18","uid___A002_X215db8_X1d5",
"uid___A002_X215db8_X392"]
for asdm in basename_all:
print "Manually removing pointing table from "+asdm+".cal.ms ... Please be careful!"
tb.open(asdm+'.cal.ms/POINTING',nomodify=False)
a = tb.rownumbers()
tb.removerows(a)
tb.close()
# In CASA
cal_south_vis = [vis+'.cal.ms' for vis in basename_south]
cal_north_vis = [vis+'.cal.ms' for vis in basename_north]
os.system('rm -rf Antennae_South.cal.ms')
concat(vis=cal_south_vis, concatvis='Antennae_South.cal.ms', timesort=T)
os.system('rm -rf Antennae_North.cal.ms')
concat(vis=cal_north_vis, concatvis='Antennae_North.cal.ms', timesort=T)
The syntax used to construct the 'cal_south_vis' variable loops over basename_south and makes a list after adding '.cal.ms' to each member. To see the list 'print cal_south_vis'.
Continue on to Imaging of the Science Target
Now you can continue on to the imaging guide.
Last checked on CASA Version 3.4.0.