NGC 5921: red-shifted HI emission 6.5.2: Difference between revisions
→Import the Data: Reformat following more modern guides |
|||
| Line 83: | Line 83: | ||
== Import the Data == | == Import the Data == | ||
The next step is to import the multisource UVFITS data to a CASA measurement set via the [https://casadocs.readthedocs.io/en/stable/api/tt/casatasks.data.importuvfits.html importuvfits] | The next step is to import the multisource UVFITS data to a CASA measurement set via the [https://casadocs.readthedocs.io/en/stable/api/tt/casatasks.data.importuvfits.html importuvfits] task. This task is used to read in data UVFITS data from the legacy VLA or other telescope; for reading in EVLA data in ASDM format the task [https://casadocs.readthedocs.io/en/v6.5.2/api/tt/casatasks.data.importasdm.html importasdm] would be used. | ||
Note that you can set each parameter for any particular task one-by-one ('interactive CASA', or you can supply the task and input parameters with a single command ('pseudo-interactive CASA'). | |||
<pre style="background-color: #d8bfd8;"> | <pre style="background-color: #d8bfd8;"> | ||
# Interactive CASA | # Interactive CASA | ||
default importuvfits | default importuvfits | ||
inp | inp | ||
fitsfile = fitsdata | fitsfile = fitsdata | ||
vis='ngc5921.demo.ms' | vis='ngc5921.demo.ms' | ||
inp | inp | ||
saveinputs('importuvfits', 'ngc5921.demo.importuvfits.saved') | saveinputs('importuvfits', 'ngc5921.demo.importuvfits.saved') | ||
go | |||
</pre> | </pre> | ||
<source lang="python"> | <source lang="python"> | ||
| Line 117: | Line 103: | ||
</source> | </source> | ||
<pre style="background-color:lightgrey;"> | |||
Where: | |||
default importuvfits # loads importuvfits with the default parameters | |||
inp # lists the inputs available for this task | |||
fitsfile='filename.fits' # sets the filename of the fits file to use | |||
vis='filename.ms' # sets the name of the output measurement set created (.ms) | |||
go # executes the task with the given inputs | |||
</pre> | |||
A few things to note: | |||
* The first [https://casadocs.readthedocs.io/en/stable/api/casashell/inp.html inp] reveals three parameters with defaults fitsfile=' '; vis=' '; antnamescheme='old'. We only set two of these, leaving antnamescheme='old' set to the default. | |||
* fitsfile=fitsdata sets the value of fitsfile to the fitsdata string variable defined earlier. If you were using your own dataset, you could enter it directly as a string (e.g., fitsfile='mydata.fits') or define it as a variable before setting fitsfile (e.g. fitsdata='mydata.fits', then fitsfile=fitsdata). You should avoid using 'fitsfile' as a variable name, as this will be overwritten by 'default importuvfits' or 'tget importuvfits'. | |||
* The second [https://casadocs.readthedocs.io/en/stable/api/casashell/inp.html inp] shows that the string variable fitsdata, defined earlier, has been expanded. A valid entry shows in blue, while an invalid entry shows in red. Default entries (as for antnamescheme) show in black. | |||
== A Summary of the Data == | == A Summary of the Data == | ||
Revision as of 18:46, 23 November 2022
This version is being edited. See NGC 5921: red-shifted HI emission for the current version of this cookbook!
Disclaimer: Due to continuous CASA software updates, GUI images may look different on more recent versions than those shown here.
Last checked on CASA Version 5.7.2.
Overview
The technique used to calibrate and image continuum datasets generally applies to spectral line observations, except that an additional calibration step is required. Bandpass calibration flattens the spectral response of the observations, ensuring that spectral channel images are properly calibrated in amplitude and phase.
The following tutorial derives from an annotated script provided in the CASA Cookbook. The script is largely reproduced and additionally annotated with figures and illustrations. It is assumed that this tutorial will be used interactively, and so interactive pauses in the original script have been removed.
DATA: The data are included with the CASA installation.
Setting up the CASA Environment
Start up CASA in the directory you want to use.
# in bash
mkdir NGC5921
cd NGC5921
casa
We'll use a python os command to get the appropriate CASA path for your installation in order to import the data. The use of os.environ.get is explained in the Appendix. This is not part of the data reduction, just an initial step to find where the example dataset has been stored when CASA was installed – you will not normally need to do this if you are working on your own data.
# In CASA
%cpaste
# Press Enter or Return, then copy/paste the following:
import os
pathname=os.environ.get('CASAPATH').split()[0]
fitsdata=pathname+'/data/demo/NGC5921.fits'
--
Scripts are of course modified and repeated to the satisfaction of observer. To help clean up the bookkeeping and further avoid issues of write privileges, remove any prior versions of the measurement set and calibration tables.
This should be done with the rmtables task, either interactively or using rmtables('table_name'), in preference to the operating system rm -rf command, as rmtables also clears data in the CASA cache.
Import the Data
The next step is to import the multisource UVFITS data to a CASA measurement set via the importuvfits task. This task is used to read in data UVFITS data from the legacy VLA or other telescope; for reading in EVLA data in ASDM format the task importasdm would be used.
Note that you can set each parameter for any particular task one-by-one ('interactive CASA', or you can supply the task and input parameters with a single command ('pseudo-interactive CASA').
# Interactive CASA
default importuvfits
inp
fitsfile = fitsdata
vis='ngc5921.demo.ms'
inp
saveinputs('importuvfits', 'ngc5921.demo.importuvfits.saved')
go
# Pseudo-interactive CASA
importuvfits(fitsfile=fitsdata,vis="ngc5921.demo.ms")
Where:
default importuvfits # loads importuvfits with the default parameters
inp # lists the inputs available for this task
fitsfile='filename.fits' # sets the filename of the fits file to use
vis='filename.ms' # sets the name of the output measurement set created (.ms)
go # executes the task with the given inputs
A few things to note:
- The first inp reveals three parameters with defaults fitsfile=' '; vis=' '; antnamescheme='old'. We only set two of these, leaving antnamescheme='old' set to the default.
- fitsfile=fitsdata sets the value of fitsfile to the fitsdata string variable defined earlier. If you were using your own dataset, you could enter it directly as a string (e.g., fitsfile='mydata.fits') or define it as a variable before setting fitsfile (e.g. fitsdata='mydata.fits', then fitsfile=fitsdata). You should avoid using 'fitsfile' as a variable name, as this will be overwritten by 'default importuvfits' or 'tget importuvfits'.
- The second inp shows that the string variable fitsdata, defined earlier, has been expanded. A valid entry shows in blue, while an invalid entry shows in red. Default entries (as for antnamescheme) show in black.
A Summary of the Data
We'll need to have a look at the observing tables to learn the calibrator and source names. The relevant command is listobs.
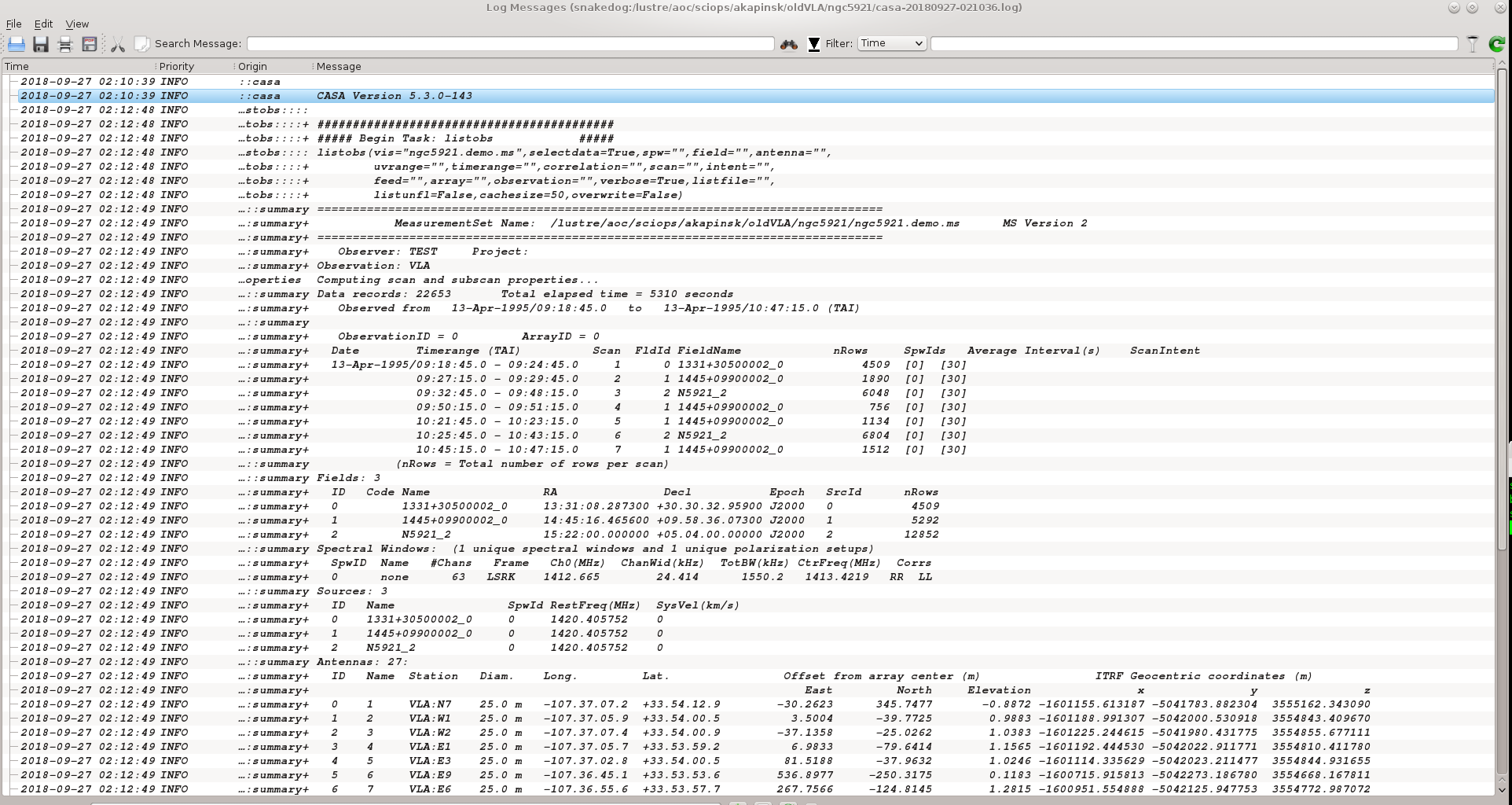
listobs(vis='ngc5921.demo.ms', verbose=True)
The output goes to the logger window; see the screenshot at right.
Tip: You can control the text size of the logger window using <ctrl>-A (smaller font) and <ctrl>-L (larger font) in Linux (<Command>-A and <Command>-L on MacOS X).
A more complete listing of the listobs output follows.
2018-09-12 20:19:48 INFO listobs ########################################## 2018-09-12 20:19:48 INFO listobs ##### Begin Task: listobs ##### 2018-09-12 20:19:48 INFO listobs listobs(vis="ngc5921.demo.ms",selectdata=True,spw="",field="",antenna="", 2018-09-12 20:19:48 INFO listobs uvrange="",timerange="",correlation="",scan="",intent="", 2018-09-12 20:19:48 INFO listobs feed="",array="",observation="",verbose=True,listfile="", 2018-09-12 20:19:48 INFO listobs listunfl=False,cachesize=50,overwrite=False) 2018-09-12 20:19:49 INFO listobs ================================================================================ 2018-09-12 20:19:49 INFO listobs MeasurementSet Name: /lustre/aoc/sciops/akapinsk/oldVLA/ngc5921/ngc5921.demo.ms MS Version 2 2018-09-12 20:19:49 INFO listobs ================================================================================ 2018-09-12 20:19:49 INFO listobs Observer: TEST Project: 2018-09-12 20:19:49 INFO listobs Observation: VLA 2018-09-12 20:19:49 INFO listobs Computing scan and subscan properties... 2018-09-12 20:19:49 INFO listobs Data records: 22653 Total elapsed time = 5310 seconds 2018-09-12 20:19:49 INFO listobs Observed from 13-Apr-1995/09:18:45.0 to 13-Apr-1995/10:47:15.0 (TAI) 2018-09-12 20:19:49 INFO listobs 2018-09-12 20:19:49 INFO listobs ObservationID = 0 ArrayID = 0 2018-09-12 20:19:49 INFO listobs Date Timerange (TAI) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent 2018-09-12 20:19:49 INFO listobs 13-Apr-1995/09:18:45.0 - 09:24:45.0 1 0 1331+30500002_0 4509 [0] [30] 2018-09-12 20:19:49 INFO listobs 09:27:15.0 - 09:29:45.0 2 1 1445+09900002_0 1890 [0] [30] 2018-09-12 20:19:49 INFO listobs 09:32:45.0 - 09:48:15.0 3 2 N5921_2 6048 [0] [30] 2018-09-12 20:19:49 INFO listobs 09:50:15.0 - 09:51:15.0 4 1 1445+09900002_0 756 [0] [30] 2018-09-12 20:19:49 INFO listobs 10:21:45.0 - 10:23:15.0 5 1 1445+09900002_0 1134 [0] [30] 2018-09-12 20:19:49 INFO listobs 10:25:45.0 - 10:43:15.0 6 2 N5921_2 6804 [0] [30] 2018-09-12 20:19:49 INFO listobs 10:45:15.0 - 10:47:15.0 7 1 1445+09900002_0 1512 [0] [30] 2018-09-12 20:19:49 INFO listobs (nRows = Total number of rows per scan) 2018-09-12 20:19:49 INFO listobs Fields: 3 2018-09-12 20:19:49 INFO listobs ID Code Name RA Decl Epoch SrcId nRows 2018-09-12 20:19:49 INFO listobs 0 1331+30500002_0 13:31:08.287300 +30.30.32.95900 J2000 0 4509 2018-09-12 20:19:49 INFO listobs 1 1445+09900002_0 14:45:16.465600 +09.58.36.07300 J2000 1 5292 2018-09-12 20:19:49 INFO listobs 2 N5921_2 15:22:00.000000 +05.04.00.00000 J2000 2 12852 2018-09-12 20:19:49 INFO listobs Spectral Windows: (1 unique spectral windows and 1 unique polarization setups) 2018-09-12 20:19:49 INFO listobs SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) Corrs 2018-09-12 20:19:49 INFO listobs 0 none 63 LSRK 1412.665 24.414 1550.2 1413.4219 RR LL 2018-09-12 20:19:49 INFO listobs Sources: 3 2018-09-12 20:19:49 INFO listobs ID Name SpwId RestFreq(MHz) SysVel(km/s) 2018-09-12 20:19:49 INFO listobs 0 1331+30500002_0 0 1420.405752 0 2018-09-12 20:19:49 INFO listobs 1 1445+09900002_0 0 1420.405752 0 2018-09-12 20:19:49 INFO listobs 2 N5921_2 0 1420.405752 0 2018-09-12 20:19:49 INFO listobs Antennas: 27: 2018-09-12 20:19:49 INFO listobs ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m) 2018-09-12 20:19:49 INFO listobs East North Elevation x y z 2018-09-12 20:19:49 INFO listobs 0 1 VLA:N7 25.0 m -107.37.07.2 +33.54.12.9 -30.2623 345.7477 -0.8872 -1601155.613187 -5041783.882304 3555162.343090 2018-09-12 20:19:49 INFO listobs 1 2 VLA:W1 25.0 m -107.37.05.9 +33.54.00.5 3.5004 -39.7725 0.9883 -1601188.991307 -5042000.530918 3554843.409670 2018-09-12 20:19:49 INFO listobs 2 3 VLA:W2 25.0 m -107.37.07.4 +33.54.00.9 -37.1358 -25.0262 1.0383 -1601225.244615 -5041980.431775 3554855.677111 2018-09-12 20:19:49 INFO listobs 3 4 VLA:E1 25.0 m -107.37.05.7 +33.53.59.2 6.9833 -79.6414 1.1565 -1601192.444530 -5042022.911771 3554810.411780 2018-09-12 20:19:49 INFO listobs 4 5 VLA:E3 25.0 m -107.37.02.8 +33.54.00.5 81.5188 -37.9632 1.0246 -1601114.335629 -5042023.211477 3554844.931655 2018-09-12 20:19:49 INFO listobs 5 6 VLA:E9 25.0 m -107.36.45.1 +33.53.53.6 536.8977 -250.3175 0.1183 -1600715.915813 -5042273.186780 3554668.167811 2018-09-12 20:19:49 INFO listobs 6 7 VLA:E6 25.0 m -107.36.55.6 +33.53.57.7 267.7566 -124.8145 1.2815 -1600951.554888 -5042125.947753 3554772.987072 2018-09-12 20:19:49 INFO listobs 7 8 VLA:W8 25.0 m -107.37.21.6 +33.53.53.0 -401.2640 -270.6305 2.2293 -1601614.059494 -5042001.699973 3554652.484758 2018-09-12 20:19:49 INFO listobs 8 9 VLA:N5 25.0 m -107.37.06.7 +33.54.08.0 -16.9948 194.1215 -0.1368 -1601168.756077 -5041869.099542 3555036.914367 2018-09-12 20:19:49 INFO listobs 9 10 VLA:W3 25.0 m -107.37.08.9 +33.54.00.1 -74.4964 -50.1921 1.1608 -1601265.132224 -5041982.597979 3554834.857504 2018-09-12 20:19:49 INFO listobs 10 11 VLA:N4 25.0 m -107.37.06.5 +33.54.06.1 -11.7487 134.3686 0.1774 -1601173.922897 -5041902.701204 3554987.495105 2018-09-12 20:19:49 INFO listobs 11 12 VLA:W5 25.0 m -107.37.13.0 +33.53.57.8 -179.2554 -120.8635 1.4872 -1601376.990711 -5041988.712764 3554776.381187 2018-09-12 20:19:49 INFO listobs 12 13 VLA:N3 25.0 m -107.37.06.3 +33.54.04.8 -8.2438 94.5297 0.3947 -1601177.362708 -5041925.112425 3554954.550128 2018-09-12 20:19:49 INFO listobs 13 14 VLA:N1 25.0 m -107.37.06.0 +33.54.01.8 -0.0030 0.0445 0.8773 -1601185.580779 -5041978.216463 3554876.396287 2018-09-12 20:19:49 INFO listobs 14 15 VLA:N2 25.0 m -107.37.06.2 +33.54.03.5 -4.7904 54.7090 0.5774 -1601180.839839 -5041947.470902 3554921.600805 2018-09-12 20:19:49 INFO listobs 15 16 VLA:E7 25.0 m -107.36.52.4 +33.53.56.5 348.8969 -162.6653 1.0336 -1600880.544215 -5042170.427468 3554741.431900 2018-09-12 20:19:49 INFO listobs 16 17 VLA:E8 25.0 m -107.36.48.9 +33.53.55.1 438.6654 -204.5038 0.5027 -1600801.910482 -5042219.412805 3554706.408864 2018-09-12 20:19:49 INFO listobs 17 18 VLA:W4 25.0 m -107.37.10.8 +33.53.59.1 -122.0163 -82.2819 1.2624 -1601315.866196 -5041985.352573 3554808.279150 2018-09-12 20:19:49 INFO listobs 18 19 VLA:E5 25.0 m -107.36.58.4 +33.53.58.8 195.8349 -91.2758 1.2155 -1601014.427180 -5042086.300814 3554800.787928 2018-09-12 20:19:49 INFO listobs 19 20 VLA:W9 25.0 m -107.37.25.1 +33.53.51.0 -491.1000 -331.2429 2.5539 -1601709.998072 -5042006.975455 3554602.355417 2018-09-12 20:19:49 INFO listobs 20 21 VLA:W6 25.0 m -107.37.15.6 +33.53.56.4 -244.9704 -165.2178 1.6861 -1601447.161927 -5041992.554228 3554739.677219 2018-09-12 20:19:49 INFO listobs 21 22 VLA:E4 25.0 m -107.37.00.8 +33.53.59.7 133.6478 -62.2829 1.0919 -1601068.773396 -5042051.970054 3554824.783566 2018-09-12 20:19:49 INFO listobs 23 24 VLA:E2 25.0 m -107.37.04.4 +33.54.01.1 40.6649 -18.9151 0.9550 -1601150.040469 -5042000.665669 3554860.702914 2018-09-12 20:19:49 INFO listobs 24 25 VLA:N6 25.0 m -107.37.06.9 +33.54.10.3 -23.2197 265.3902 -0.4819 -1601162.569974 -5041829.054708 3555095.873969 2018-09-12 20:19:49 INFO listobs 25 26 VLA:N9 25.0 m -107.37.07.8 +33.54.19.0 -46.5533 532.1581 -1.8550 -1601139.422904 -5041679.082136 3555316.518142 2018-09-12 20:19:49 INFO listobs 26 27 VLA:N8 25.0 m -107.37.07.5 +33.54.15.8 -38.0437 434.7201 -1.3387 -1601147.894127 -5041733.868915 3555235.935926 2018-09-12 20:19:49 INFO listobs 27 28 VLA:W7 25.0 m -107.37.18.4 +33.53.54.8 -319.1171 -215.2368 1.9407 -1601526.340031 -5041996.897001 3554698.302182 2018-09-12 20:19:49 INFO listobs ##### End Task: listobs ##### 2018-09-12 20:19:49 INFO listobs ##########################################
Key Information from listobs
Certainly the output of listobs is dense with information, but there are some particularly vital data that we'll need for the calibration.
- The calibrators are 1331+305* (3C286, the flux and bandpass calibrator) and 1445+099* (the phase calibrator). We can use wild-cards 1331* and 1445* since they uniquely identify the sources.
- The calibrator field indices are field='0' (1331+305) and field='1' (1445+099).
- The name of the source in the observations list is N5921_2, or field = '2'.
- The data were taken in a single IF (a single spectral window, SpwID = 0), divided into 63 channels.
- Only RR and LL correlations are present; cross-pols are absent.
Flagging
Flag the autocorrelations
We don't need the autocorrelation data, and we can use flagdata to get rid of them. You shouldn't have to specify the measurement set, because the variable vis is already set, but it never hurts to be cautious.
flagdata(vis="ngc5921.demo.ms", autocorr=True)
CASA issues a warning when running this command, but this can be disregarded.
Interactive Flagging
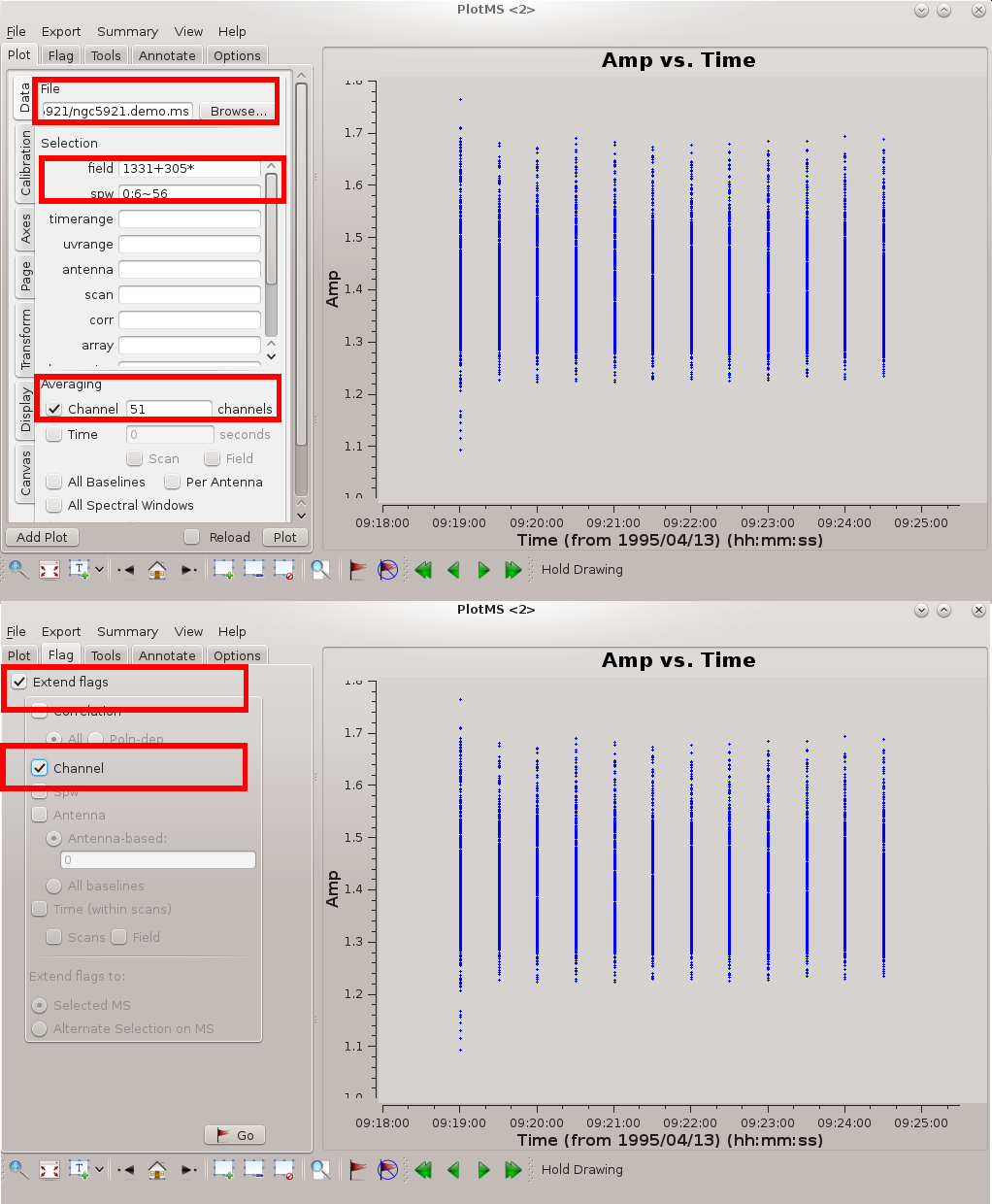
plotms is a good tool for flagging spectral line data. Check out the tutorial that describes editing VLA continuum data. Spectral line data of course require some consideration of channels and channel averaging.
plotms()
The figure at right highlights the settings needed for effective editing of a spectral line data set. The key settings are as follows.
- Specify the measurement set in File; the Browse button allows you to hunt down the measurement set.
- It's better to edit one source at a time. In the illustrated example, the flux / bandpass calibrator 1331+305* is displayed.
- Average the channels. First, specify the central channels to remove band edge effects. Channels 6~56 in the first spectral window (IF) are appropriate (see #Inspect the Bandpass Response Curve, below). In plotms() specify: spw=0:6~56. In the Channel Averaging box, enter 51 channels to average over all channels in the given range.
- Ideally you want the channels to have the same (u, v) coverage (projected baseline spacings as viewed from the source); otherwise, the beam (point spread function) will be different for each channel. Therefore, if you flag data from a given channel it's usually a good idea to flag those data from all channels. Under the Flagging tab, specify Extend flags to Channel.
With these settings, interactive flagging proceeds as for continuum data. When you're satisfied with the edits, File → Quit to return to the CASA prompt.
Calibration
Calibration of spectral line data broadly follows the approach for continuum data, except that the amplitude and phase corrections are a function of frequency and so must be corrected by bandpass calibration. The basic calibration steps follow.
- Set the flux scale of the primary calibrator, here, 1331+305 = 3C 286.
- Determine bandpass corrections based on the primary calibrator. In the script that follows, the bandpass calibration will be stored in ngc5921.demo.bcal.
- Inspect the bandpass correction to determine viable channels for averaging and imaging. We want to toss out end channels where the response is poor.
- Determine the gain calibrations on the bandpass-corrected and channel-averaged data. In this step, we effectively turn the spectral line data into a single-channel continuum data set and calibrate accordingly. The calibration is first stored in ngc5921.demo.gcal. In the second part of this step we correct the fluxscale of the .gcal table, and store the final calibration solutions with correct fluxes in ngc5921.demo.fluxscale (this is the table that needs to be applied later to the data, not the .gcal version).
- Inspect the gain calibration solutions to look for any aberrant solutions that hint at bad calibrator data.
- Apply the calibration solutions to the source (N5921_2). This action literally adds a new column of data to the measurement set. This new column contains the data with the gain calibration and bandpass calibration applied, but it does not overwrite the raw data in case the calibration needs revision.
Setting the Flux Scale
setjy generates a source model for the primary calibrator, 1331+305 = 3C286. From CASA 5.3+ the default standard is Perley-Butler 2017, and includes resolved structure of the calibrators. This is 1.4GHz D-config and 1331+305 is sufficiently unresolved that, in principle, we don't need a model image; however, here we proceed with applying the detailed model, as a good practice.
setjy also looks up the radio SED for common flux calibrators and automatically assigns the total flux density.
# 1331+305 = 3C286 is our primary calibrator. Use the wildcard on the end of the source name
setjy(vis='ngc5921.demo.ms', field='1331+305*', model='3C286_L.im')
A summary of the operation is sent to the logger window. Here's a listing of the output.
2018-09-12 20:43:38 INFO setjy ##########################################
2018-09-12 20:43:38 INFO setjy ##### Begin Task: setjy #####
2018-09-12 20:43:38 INFO setjy setjy(vis="ngc5921.demo.ms",field="1331+305*",spw="",selectdata=False,timerange="",
2018-09-12 20:43:38 INFO setjy scan="",intent="",observation="",scalebychan=True,standard="Perley-Butler 2017",
2018-09-12 20:43:38 INFO setjy model="3C286_L.im",modimage="",listmodels=False,fluxdensity=-1,spix=0.0,
2018-09-12 20:43:38 INFO setjy reffreq="1GHz",polindex=[],polangle=[],rotmeas=0.0,fluxdict={},
2018-09-12 20:43:38 INFO setjy useephemdir=False,interpolation="nearest",usescratch=False,ismms=False)
2018-09-12 20:43:38 INFO setjy {'field': '1331+305*'}
2018-09-12 20:43:38 INFO Imager Opening MeasurementSet [...]
2018-09-12 20:43:38 INFO setjy Using /home/casa/data/distro/nrao/VLA/CalModels/3C286_L.im for modimage.
2018-09-12 20:43:38 INFO setjy CASA Version 5.3.0-143
2018-09-12 20:43:38 INFO setjy
2018-09-12 20:43:39 INFO imager Using channel dependent flux densities
2018-09-12 20:43:39 INFO imager Selected 4509 out of 22653 rows.
2018-09-12 20:43:39 INFO imager 1331+30500002_0 (fld ind 0) spw 0 [I=15.016, Q=0, U=0, V=0] Jy @ 1.4127e+09Hz, (Perley-Butler 2017)
2018-09-12 20:43:40 INFO imager Using model image /home/casa/data/distro/nrao/VLA/CalModels/3C286_L.im
2018-09-12 20:43:40 INFO imager Scaling spw(s) [0]'s model image by channel to I = 15.0159, 15.0118, 15.0077 Jy @(1.41265e+09, 1.41343e+09, 1.41419e+09)Hz (LSRK) for visibility prediction (a few representative values are shown).
2018-09-12 20:43:40 INFO imager The model image's reference pixel is 0.00904522 arcsec from 1331+30500002_0's phase center.
2018-09-12 20:43:40 INFO imager Will clear any existing model with matching field=1331+30500002_0 and spw=*
2018-09-12 20:43:40 INFO Clearing model records in MS header for selected fields.
2018-09-12 20:43:40 INFO 1331+30500002_0 (id = 0) deleted.
2018-09-12 20:43:40 INFO imager Selected 4509 out of 22653 rows.
2018-09-12 20:43:40 INFO setjy ##### End Task: setjy #####
2018-09-12 20:43:40 INFO setjy ##########################################
Bandpass Calibration
The flux calibrator 1331+305 = 3C 286 now has a model assigned to it. Since the bandwidth of our observations is only 1.55 MHz, the model doesn't change over this narrow range of frequencies, so we can use it to determine amplitude and phase (gain) corrections for each channel independently. The result is the bandpass calibration.
As for any antenna-based calibration scheme, we have to pick an antenna to act as the reference point for the calibration. Any antenna will do, but it's better to pick one near the center of the array. For the remainder of the calibration, we will use refant = '15'.
# We can first do the bandpass on the single 5min scan on 1331+305. At 1.4GHz phase stablility should be sufficient to do this without
# a first (rough) gain calibration. This will give us the relative antenna gain as a function of frequency.
bandpass(vis='ngc5921.demo.ms', caltable='ngc5921.demo.bcal', field='0', selectdata=False, bandtype='B', solint='inf', combine='scan', refant='15')
- field='0' : Use the flux calibrator 1331+305 = 3C286 (FIELD_ID 0) as bandpass calibrator.
- bandtype='B' : Choose bandpass solution type. Pick standard time-binned B (rather than BPOLY).
- solint='inf' and combine='scan' : Set solution interval arbitrarily long (get single bandpass).
- refant = '15' : Reference antenna Name 15 (15=VLA:N2) (Id 14)
Inspect the Bandpass Response Curve
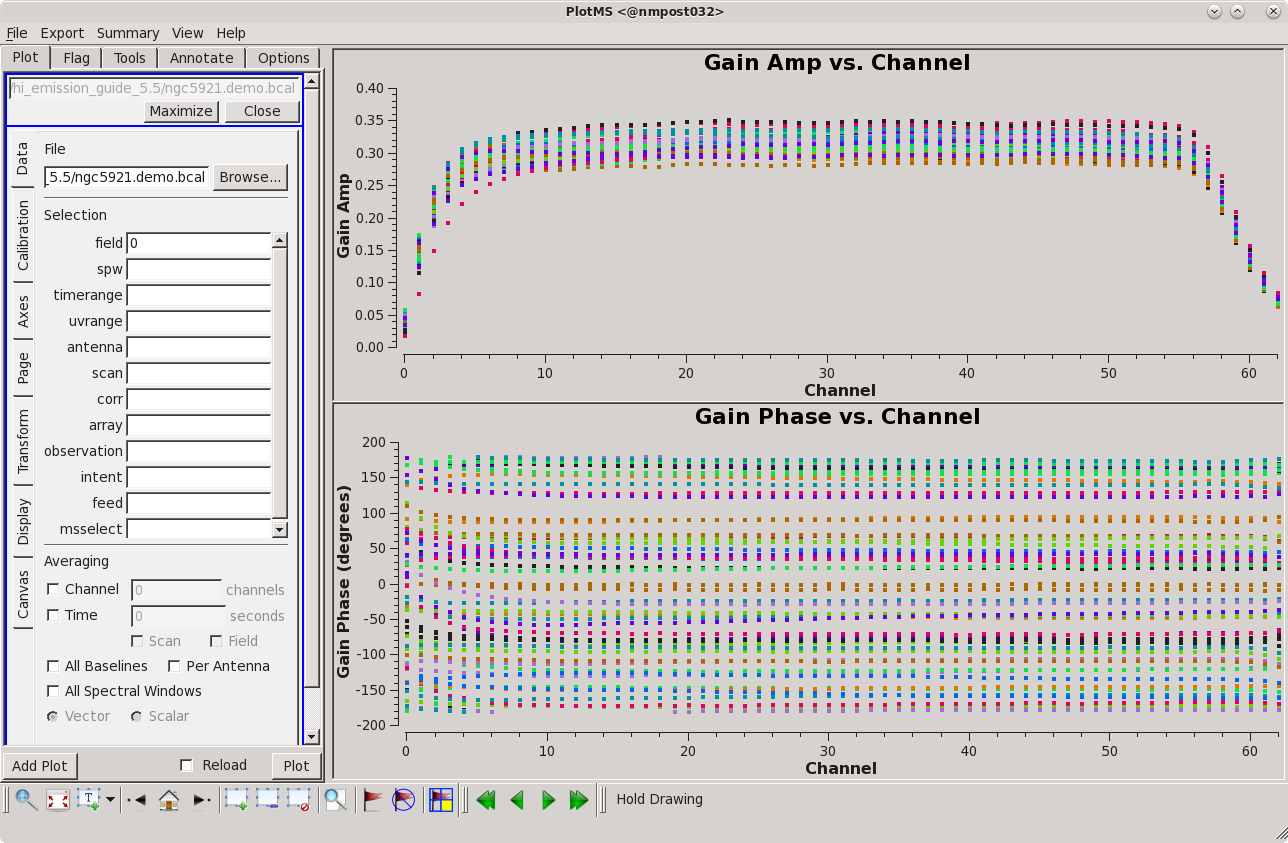
In the gain calibration to follow, we will effectively convert the spectral line data into a continuum data set. Before proceeding, we need to inspect the bandpass calibration to make sure that it contains no bad values and also to inspect which channels to average to produce the continuum data. Plotms is the standard tool for plotting calibration solutions. The following commands produce the figure at right.
# Set up 2x1 panels - upper panel amp vs. channel
plotms(vis='ngc5921.demo.bcal', field='0', gridrows=2, gridcols=1, plotindex=0, rowindex=0, xaxis='channel', yaxis='amp', showgui=True, clearplots=False, coloraxis='antenna1')
# Set up 2x1 panels - lower panel phase vs. channel
plotms(vis='ngc5921.demo.bcal', field='0', gridrows=2, gridcols=1, plotindex=1, rowindex=1, xaxis='channel', yaxis='phase', showgui=True, clearplots=False, coloraxis='antenna1')
By inspection, the amplitude response curve is flat over channels 6~56; that channel range will be used to generate the continuum data for gain calibration. If you want to further inspect the plots interactively and iterate over antenna, set iteraxis = 'antenna'
Notice that plotms is run twice: once to display gain amplitudes as a function of channel (frequency), and again to plot gain phases as a function of channel.
Gain Calibration
From inspection of the bandpass response curve, we can average channels 6~56 to produce continuum data for the calibrators. For VLA data, this averaging is specified through the spw (spectral window) parameter, which takes the form IF:Channel-range, as follows.
spw = '0:6~56'
That is, there is only one spectral window (IF), spw = 0, and we want to average channels 6~56 within that spectral window.
Gain calibrations are otherwise determined as for continuum data.
- gaincal() is run only on the calibrators, 1331+305 (flux calibrator) and 1445+099 (phase calibrator).
- The default model for gain calibrations is a 1 Jy point-source. The flux scale is overridden by setjy, which has been performed for the flux calibrator. We need to transfer that flux scale to the phase calibrator using fluxscale().
- Note that fluxscale() determines the flux density of the phase calibrator and accordingly adjusts its model and calibration solutions. A report of the results are sent to the logger window.
- Unless you use parameter incremental=True while executing fluxscale() (the default is False), the resulting .fluxscale table will replace the .gcal table at this point. This particularly important in the applycal() stage.
# Armed with the bandpass, we now solve for the time-dependent antenna gains using our previously determined bandpass.
# Note this will automatically be applied to all sources not just the one used to determine the bandpass
gaincal(vis='ngc5921.demo.ms', caltable='ngc5921.demo.gcal', gaintable=['ngc5921.demo.bcal'], interp=['nearest'], field='0,1', spw='0:6~56', gaintype='G', solint='inf', calmode='ap', refant='15')
# Now we will transfer the flux scale to the phase calibrator.
# We will be using 1331+305 (the source we did setjy on) as our flux standard reference.
# Note its extended name as in the FIELD table summary above (it has a VLA seq number appended)
fluxscale(vis='ngc5921.demo.ms', fluxtable='ngc5921.demo.fluxscale', caltable='ngc5921.demo.gcal', reference='1331*', transfer='1445*')
The output from fluxscale follows. A relatively large uncertainty for the phase calibrator is a sign that something went wrong, perhaps bad solutions in gaincal. Here, the phase calibrator scaled to 2.486 ± 0.001 Jy, which looks reasonable.
2018-09-12 22:38:05 INFO fluxscale ########################################## 2018-09-12 22:38:05 INFO fluxscale ##### Begin Task: fluxscale ##### 2018-09-12 22:38:05 INFO fluxscale fluxscale(vis="ngc5921.demo.ms",caltable="ngc5921.demo.gcal",fluxtable="ngc5921.demo.fluxscale",reference="1331*",transfer="1445*", 2018-09-12 22:38:05 INFO fluxscale listfile="",append=False,refspwmap=[-1],gainthreshold=-1.0,antenna="", 2018-09-12 22:38:05 INFO fluxscale timerange="",scan="",incremental=False,fitorder=1,display=False) 2018-09-12 22:38:05 INFO fluxscale ****Using NEW VI2-driven calibrater tool**** 2018-09-12 22:38:05 INFO fluxscale Opening MS: ngc5921.demo.ms for calibration. 2018-09-12 22:38:05 INFO fluxscale Initializing nominal selection to the whole MS. 2018-09-12 22:38:05 INFO fluxscale Beginning fluxscale--(MSSelection version)------- 2018-09-12 22:38:05 INFO fluxscale Found reference field(s): 1331+30500002_0 2018-09-12 22:38:05 INFO fluxscale Found transfer field(s): 1445+09900002_0 2018-09-12 22:38:05 INFO fluxscale Flux density for 1445+09900002_0 in SpW=0 (freq=1.41342e+09 Hz) is: 2.52609 +/- 0.00218206 (SNR = 1157.67, N = 54) 2018-09-12 22:38:05 INFO fluxscale Storing result in ngc5921.demo.fluxscale 2018-09-12 22:38:05 INFO fluxscale Writing solutions to table: ngc5921.demo.fluxscale 2018-09-12 22:38:06 INFO fluxscale CASA Version 5.3.0-143 2018-09-12 22:38:06 INFO fluxscale 2018-09-12 22:38:07 INFO fluxscale ##### End Task: fluxscale ##### 2018-09-12 22:38:07 INFO fluxscale ##########################################
Inspect the Calibration Solutions
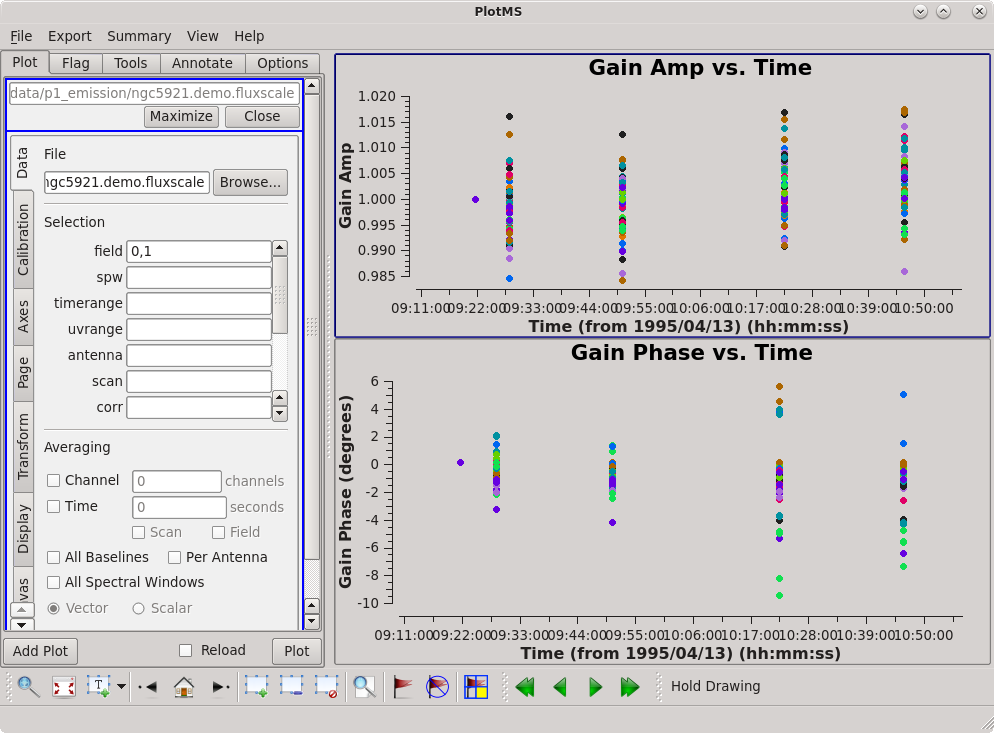
Now inspect the results of gaincal. The setup is identical to that used to plot the bandpass response curve. The only change is that we are plotting the gaintable ngc5921.demo.gcal, and we're looking at solutions for both of the calibrator sources. The results are shown at right.
# Set up 2x1 panels - upper panel amp vs. time
plotms(vis='ngc5921.demo.fluxscale', field='0,1', gridrows=2, gridcols=1, plotindex=0, rowindex=0, yaxis='amp', showgui=True, clearplots=False, coloraxis='antenna1')
# Set up 2x1 panels - lower panel phase vs. time
plotms(vis='ngc5921.demo.fluxscale', field='0,1', gridrows=2, gridcols=1, plotindex=1, rowindex=1, yaxis='phase', showgui=True, clearplots=False, coloraxis='antenna1')
The amp and phase coherence looks good. If you want to do this interactively and iterate over antenna, set iteraxis = 'antenna'.
Apply the Solutions
Next, apply the calibration solutions to the calibrators themselves, and finally transfer the calibration solutions by interpolation (or nearest-neighbor sampling) to the source. The relevant task is applycal, which fills out a new column (CORRECTED_DATA) of calibrated data in the measurement set without wiping out the raw data column. The application is identical to that used for continuum data, except that the bandpass table is also included in the calibration. To apply multiple calibrations at once, provide the gaintable parameter with a list of calibration tables, as follows.
gaintable = ['ngc5921.demo.fluxscale', 'ngc5921.demo.bcal']
We want to correct the calibrators using themselves and transfer from 1445+099 to itself and the target N5921. Start with the fluxscale/gain and bandpass tables. We will pick the 1445+099 out of the gain table for transfer and use all of the bandpass table. Also, note that the table .fluxscale has the .gcal solutions with the correct flux scale applied, and so there is no need to invoke the .gcal again in the applycal() command below.
applycal(vis='ngc5921.demo.ms', field='1,2', gaintable=['ngc5921.demo.fluxscale','ngc5921.demo.bcal'], gainfield=['1','*'],
interp=['linear','nearest'], spwmap=[], selectdata=False)
Now for completeness apply 1331+305 to itself.
applycal(vis='ngc5921.demo.ms', field='0', gaintable=['ngc5921.demo.fluxscale','ngc5921.demo.bcal'], gainfield=['0','*'],
interp=['linear','nearest'], spwmap=[], selectdata=False)
Plot the Spectrum
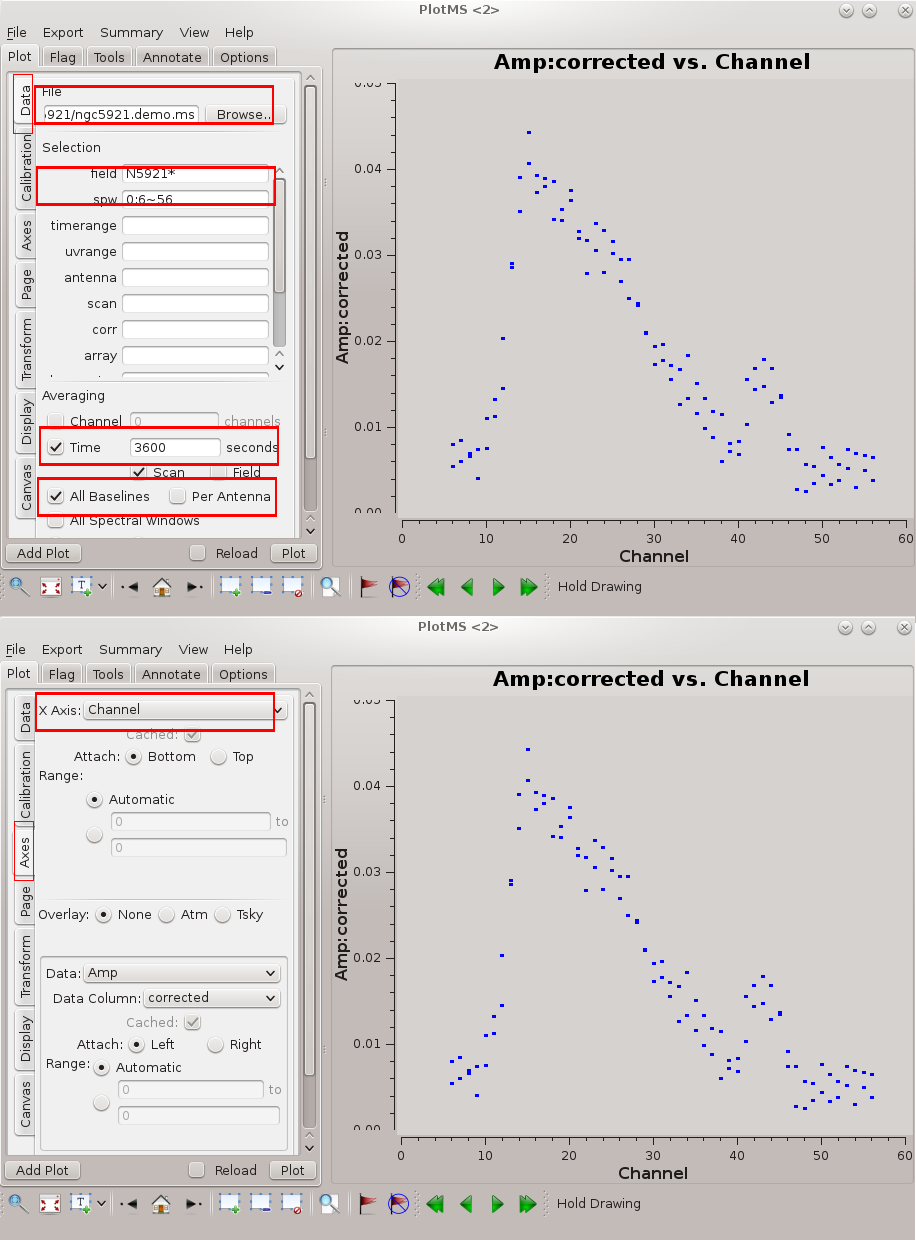
Before we attempt to image the 21 cm cube of the source, we need to subtract off the underlying continuum, which means we need to plot the integrated spectrum of the source to determine the continuum channels.
We can do this in plotms.
plotms(vis='ngc5921.demo.ms', selectdata=True, field='N5921*', spw='0:6~56', averagedata=True, avgtime='3600', avgscan=True,
avgbaseline=True, xaxis='channel', yaxis='amp', ydatacolumn='corrected')
Note that we have entered all the relevent parameters via the task interface, as an alternative to entering each option into the GUI. If the symbols appear too small, the size may be increased via the Display tab: change the Unflagged Points Symbol to 'Custom' and increase the number of pixels for the plotting symbol. The resulting plot is illustrated in the figure at right. Briefly, we want to average both in time and over baselines to get the signal-to-noise necessary to reveal the 21 cm profile (see Averaging data in plotms for more details on averaging options). If you wish to enter the values directly into the GUI, you can follow the (Tab)Command convention of the flagging tutorial with the following settings :
- (Data)field = N5921*
- (Data)spw = 0:6~56
- (Data)Averaging → Time = 3600 (average over some long time window)
- (Data)Averaging → Scan = True (checkmark; average in time across scan boundaries)
- (Data)Averaging → All Baselines = True (checkmark)
- (Axes)X Axis = Channel
- (Axes)Y Axis = Amp
Now remove 0:6~56 from the spw field to see all channels. From inspection of this plot, it looks like channels 4~6 and 50~59 contain line-free channels, suitable to use for continuum subtraction.
Continuum Subtraction
The next step is to split off the NGC 5921 data from the multisource measurement set and subtract the continuum. Splitting uses the split command, as follows.
split(vis='ngc5921.demo.ms', outputvis='ngc5921.demo.src.split.ms', field='N5921*', spw='', datacolumn='corrected')
This action generated a new measurement set called ngc5921.demo.src.split.ms and copied the calibrated source data (datacolumn = 'corrected') into it.
uvcontsub subtracts the continuum from the data in the visibility (u, v) plane. We will be using channels 4-6 and 50-59 for continuum.
uvcontsub(vis='ngc5921.demo.src.split.ms', field='N5921*', fitspw='0:4~6;50~59', spw='0', solint=0.0, fitorder=0, want_cont=True)
Notice that uvcontsub splits the data into two new measurement sets, 'ngc5921.demo.ms.cont', which contains an average of the continuum channels, and 'ngc5921.demo.ms.contsub', which contains the continuum-subtracted spectral line data.
Imaging
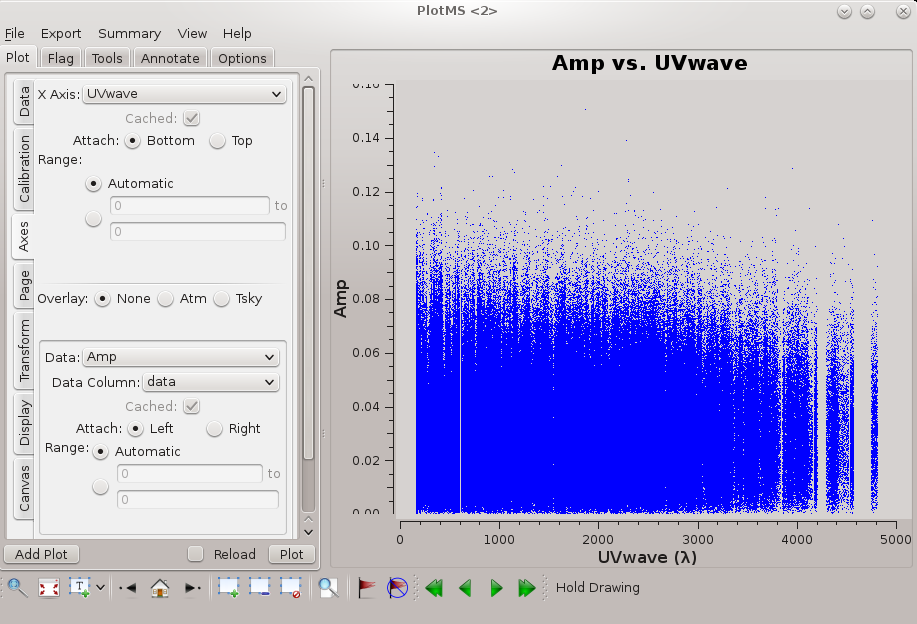
Now we can generate the primary science product, a tclean data cube (ra, dec, velocity) from the continuum-subtracted (u, v, channel) measurement set, ngc5921.demo.ms.contsub. Things to consider in using tclean:
- To ensure channels aren't averaged prior to imaging, choose mode='channel'.
- Specify the channels to image using start = 5, width = 1 (no averaging over channels), nchan = 46; only channels 5~51 will be imaged.
- The maximum baseline is just under 5 kilolambda (see the figure at right), and so the expected synthetic beam is roughly 1.22 × 206265 / 5000 = 50 arcseconds (subject to the details of u, v weighting). Pixels should sample the beam better than 3 times, so 15 arcseconds is a good choice of pixel size (cell = ['15.0arcsec','15.0arcsec']).
- We only want to tclean down to the noise, which is easily determined by trial-and-error imaging of a single channel (choosing nchan=1 and start appropriately). Here, tclean stops when the maximum residual on the channel is below threshold='8.0mJy'.
# Image the continuum subtracted measurement set
tclean(vis='ngc5921.demo.src.split.ms.contsub', imagename='ngc5921.demo.tclean', field='0', datacolumn='data', specmode='cube', nchan=46, start=5, width=1, spw='', deconvolver='hogbom', gridder='standard', niter=6000, gain=0.1, threshold='8.0mJy', imsize=[256,256], cell=['15.0arcsec','15.0arcsec'], weighting='briggs', robust=0.5, mask = 'box[[108pix,108pix],[148pix,148pix]]', interactive=False, pblimit=-0.2)
Use imhead to look at the cube header:
imhead(imagename='ngc5921.demo.tclean.image', mode='summary')
The output, as follows, appears in the logger window.
2019-09-03 22:25:25 INFO imhead ##### Begin Task: imhead ##### 2019-09-03 22:25:25 INFO imhead imhead(imagename="ngc5921.demo.tclean.image",mode="summary",hdkey="",hdvalue="",verbose=False) 2019-09-03 22:25:25 INFO ImageMetaData 2019-09-03 22:25:25 INFO ImageMetaData Image name : ngc5921.demo.tclean.image 2019-09-03 22:25:25 INFO ImageMetaData Object name : N5921_2 2019-09-03 22:25:25 INFO ImageMetaData Image type : PagedImage 2019-09-03 22:25:25 INFO ImageMetaData Image quantity : Intensity 2019-09-03 22:25:25 INFO ImageMetaData Pixel mask(s) : None 2019-09-03 22:25:25 INFO ImageMetaData Region(s) : None 2019-09-03 22:25:25 INFO ImageMetaData Image units : Jy/beam 2019-09-03 22:25:25 INFO ImageMetaData Restoring Beams 2019-09-03 22:25:25 INFO ImageMetaData Pol Type Chan Freq Vel 2019-09-03 22:25:25 INFO ImageMetaData I Max 7 1.41296e+09 1571.92 51.6639 arcsec x 47.3594 arcsec pa= 8.3205 deg 2019-09-03 22:25:25 INFO ImageMetaData I Min 43 1.41384e+09 1386.42 51.5897 arcsec x 47.3127 arcsec pa= 8.3869 deg 2019-09-03 22:25:25 INFO ImageMetaData I Median 13 1.41310e+09 1541.00 51.6611 arcsec x 47.3318 arcsec pa= 8.1425 deg 2019-09-03 22:25:25 INFO ImageMetaData 2019-09-03 22:25:25 INFO ImageMetaData Direction reference : J2000 2019-09-03 22:25:25 INFO ImageMetaData Spectral reference : LSRK 2019-09-03 22:25:25 INFO ImageMetaData Velocity type : RADIO 2019-09-03 22:25:25 INFO ImageMetaData Rest frequency : 1.42041e+09 Hz 2019-09-03 22:25:25 INFO ImageMetaData Pointing center : 15:22:00.000000 +05.04.00.000000 2019-09-03 22:25:25 INFO ImageMetaData Telescope : VLA 2019-09-03 22:25:25 INFO ImageMetaData Observer : TEST 2019-09-03 22:25:25 INFO ImageMetaData Date observation : 1995/04/13/09:33:00 2019-09-03 22:25:25 INFO ImageMetaData Telescope position: [-1.60119e+06m, -5.04198e+06m, 3.55488e+06m] (ITRF) 2019-09-03 22:25:25 INFO ImageMetaData 2019-09-03 22:25:25 INFO ImageMetaData Axis Coord Type Name Proj Shape Tile Coord value at pixel Coord incr Units 2019-09-03 22:25:25 INFO ImageMetaData ------------------------------------------------------------------------------------------------ 2019-09-03 22:25:25 INFO ImageMetaData 0 0 Direction Right Ascension SIN 256 64 15:22:00.000 128.00 -1.500000e+01 arcsec 2019-09-03 22:25:25 INFO ImageMetaData 1 0 Direction Declination SIN 256 64 +05.04.00.000 128.00 1.500000e+01 arcsec 2019-09-03 22:25:25 INFO ImageMetaData 2 1 Stokes Stokes 1 1 I 2019-09-03 22:25:25 INFO ImageMetaData 3 2 Spectral Frequency 46 8 1.41279e+09 0.00 2.4414062e+04 Hz 2019-09-03 22:25:25 INFO ImageMetaData Velocity 1607.99 0.00 -5.152860e+00 km/s 2019-09-03 22:25:25 INFO imhead ##### End Task: imhead ##### 2019-09-03 22:25:25 INFO imhead ##########################################
Additional Science Products
If things went well, you should now have a spectral line cube (ngc5921.demo.tclean.image) as a primary science product. The demo script illustrates further how to generate cube statistics (using imstat), an integrated spectrum, and moment maps.
Cube Statistics
imstat is the tool for displaying statistics of images and cubes. The following example displays the statistics for an empty region of the whole cube.
cubestat=imstat(imagename='ngc5921.demo.tclean.image', box='10,10,100,100')
2019-09-03 22:40:15 INFO imstat ########################################## 2019-09-03 22:40:15 INFO imstat ##### Begin Task: imstat ##### 2019-09-03 22:40:15 INFO imstat imstat(imagename="ngc5921.demo.tclean.image",axes=-1,region="",box="10,10,100,100",chans="", 2019-09-03 22:40:15 INFO imstat stokes="",listit=True,verbose=True,mask="",stretch=False, 2019-09-03 22:40:15 INFO imstat logfile="",append=True,algorithm="classic",fence=-1,center="mean", 2019-09-03 22:40:15 INFO imstat lside=True,zscore=-1,maxiter=-1,clmethod="auto",niter=3) 2019-09-03 22:40:15 INFO imstat Using specified box(es) 10,10,100,100 2019-09-03 22:40:15 INFO imstat Determining stats for image ngc5921.demo.tclean.image 2019-09-03 22:40:15 INFO imstat Selected bounding box : 2019-09-03 22:40:15 INFO imstat [10, 10, 0, 0] to [100, 100, 0, 45] (15:23:58.379, +04.34.29.305, I, 1.41279e+09Hz to 15:22:28.105, +04.56.59.962, I, 1.41389e+09Hz) 2019-09-03 22:40:15 INFO imstat Statistics calculated using Classic algorithm 2019-09-03 22:40:15 INFO imstat Regions --- 2019-09-03 22:40:15 INFO imstat -- bottom-left corner (pixel) [blc]: [10, 10, 0, 0] 2019-09-03 22:40:15 INFO imstat -- top-right corner (pixel) [trc]: [100, 100, 0, 45] 2019-09-03 22:40:15 INFO imstat -- bottom-left corner (world) [blcf]: 15:23:58.379, +04.34.29.305, I, 1.41279e+09Hz 2019-09-03 22:40:15 INFO imstat -- top-right corner (world) [trcf]: 15:22:28.105, +04.56.59.962, I, 1.41389e+09Hz 2019-09-03 22:40:15 INFO imstat Values --- 2019-09-03 22:40:15 INFO imstat -- flux [flux]: 1.99425 Jy.km/s 2019-09-03 22:40:15 INFO imstat -- number of points [npts]: 380926 2019-09-03 22:40:15 INFO imstat -- maximum value [max]: 0.00953276 Jy/beam 2019-09-03 22:40:15 INFO imstat -- minimum value [min]: -0.0100478 Jy/beam 2019-09-03 22:40:15 INFO imstat -- position of max value (pixel) [maxpos]: [85, 63, 0, 8] 2019-09-03 22:40:15 INFO imstat -- position of min value (pixel) [minpos]: [30, 18, 0, 7] 2019-09-03 22:40:15 INFO imstat -- position of max value (world) [maxposf]: 15:22:43.151, +04.47.44.907, I, 1.41298e+09Hz 2019-09-03 22:40:15 INFO imstat -- position of min value (world) [minposf]: 15:23:38.319, +04.36.29.518, I, 1.41296e+09Hz 2019-09-03 22:40:15 INFO imstat -- Sum of pixel values [sum]: 4.76561 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Sum of squared pixel values [sumsq]: 1.38125 Jy/beam.Jy/beam 2019-09-03 22:40:15 INFO imstat Statistics --- 2019-09-03 22:40:15 INFO imstat -- Mean of the pixel values [mean]: 1.25106e-05 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Variance of the pixel values : 3.62589e-06 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Standard deviation of the Mean [sigma]: 0.00190418 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Root mean square [rms]: 0.00190421 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Median of the pixel values [median]: 6.37541e-06 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Median of the deviations [medabsdevmed]: 0.00127676 Jy/beam 2019-09-03 22:40:15 INFO imstat -- IQR [quartile]: 0.00255348 Jy/beam 2019-09-03 22:40:15 INFO imstat -- First quartile [q1]: -0.00126604 Jy/beam 2019-09-03 22:40:15 INFO imstat -- Third quartile [q3]: 0.00128744 Jy/beam 2019-09-03 22:40:15 INFO imstat Sum column unit = Jy/beam 2019-09-03 22:40:15 INFO imstat Mean column unit = Jy/beam 2019-09-03 22:40:15 INFO imstat Std_dev column unit = Jy/beam 2019-09-03 22:40:15 INFO imstat Minimum column unit = Jy/beam 2019-09-03 22:40:15 INFO imstat Maximum column unit = Jy/beam 2019-09-03 22:40:15 INFO imstat Npts Sum Mean Rms Std_dev Minimum Maximum 2019-09-03 22:40:15 INFO imstat 3.809260e+05 4.765615e+00 1.251060e-05 1.904215e-03 1.904176e-03 -1.004781e-02 9.532760e-03 2019-09-03 22:40:15 INFO imstat ##### End Task: imstat ##### 2019-09-03 22:40:15 INFO imstat ##########################################
The Integrated Spectrum
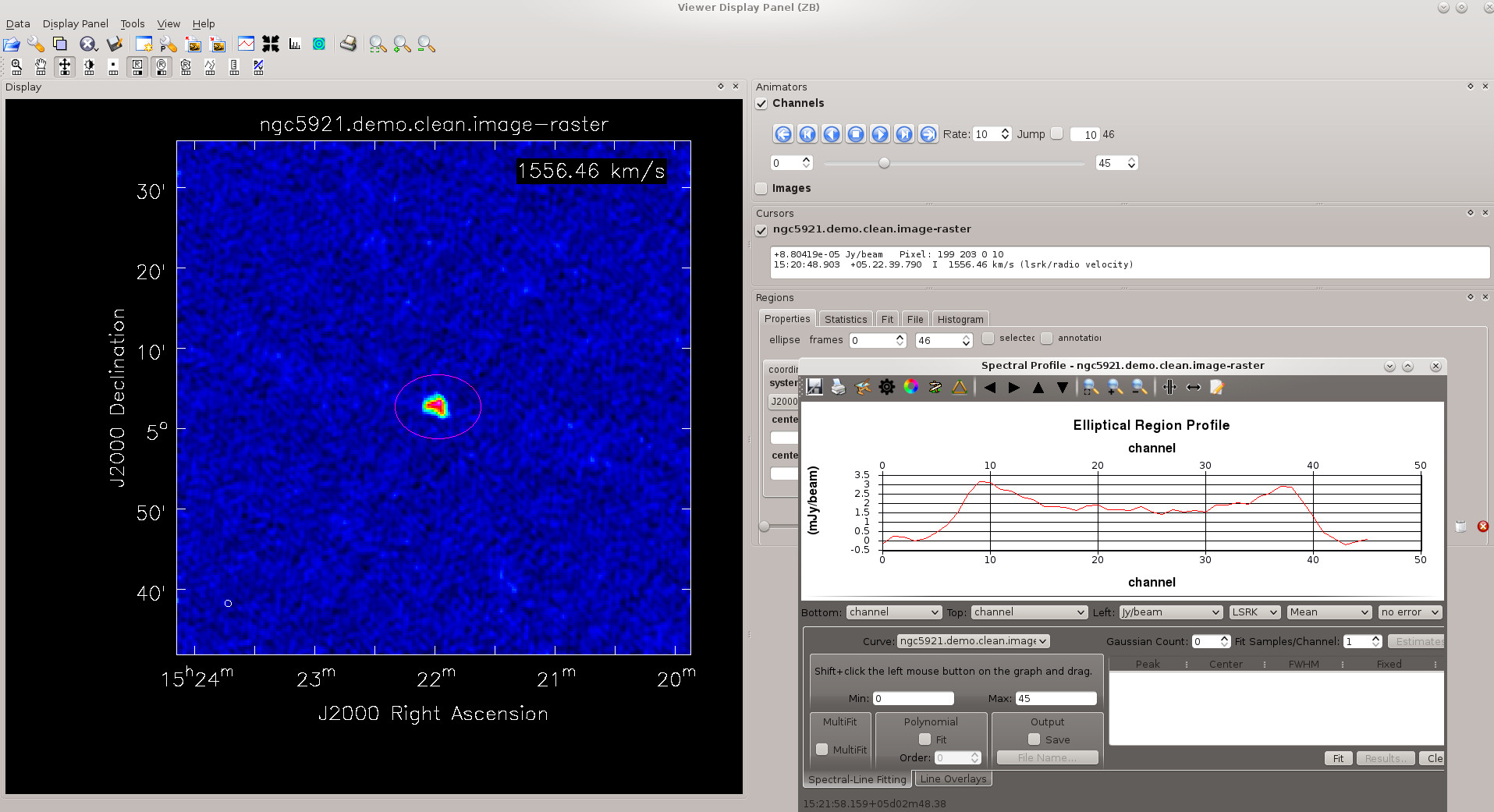
We saw earlier how to generate an integrated spectrum from the (u, v) measurement set. Here's how to produce the integrated spectrum from the spectral line cube. First, load the cube into viewer.
viewer(infile='ngc5921.demo.tclean.image')
To generate the integrated spectrum, perform the following tasks.
- Use the player controls
 to inspect the cube one channel at a time.
to inspect the cube one channel at a time. - From the viewer Tools menu, select Spectral Profile. A new graphics window should appear.
- By default, the rectangle selection tool
 is assigned to the right mouse button, and you can just right-click and drag a box over the region where you want to (spatially) integrate the spectrum. See the figure at upper right.
is assigned to the right mouse button, and you can just right-click and drag a box over the region where you want to (spatially) integrate the spectrum. See the figure at upper right. - Alternatively, you can assign one of the other selection tools by right-clicking on the appropriate button.
- The spectrum now appears in the graphics window; see the figure at right.
You can save the integrated spectrum to a text file by clicking the ![]() button on the graphics window. There are also buttons to print the figure or save the figure to disk.
button on the graphics window. There are also buttons to print the figure or save the figure to disk.
Cube Moments
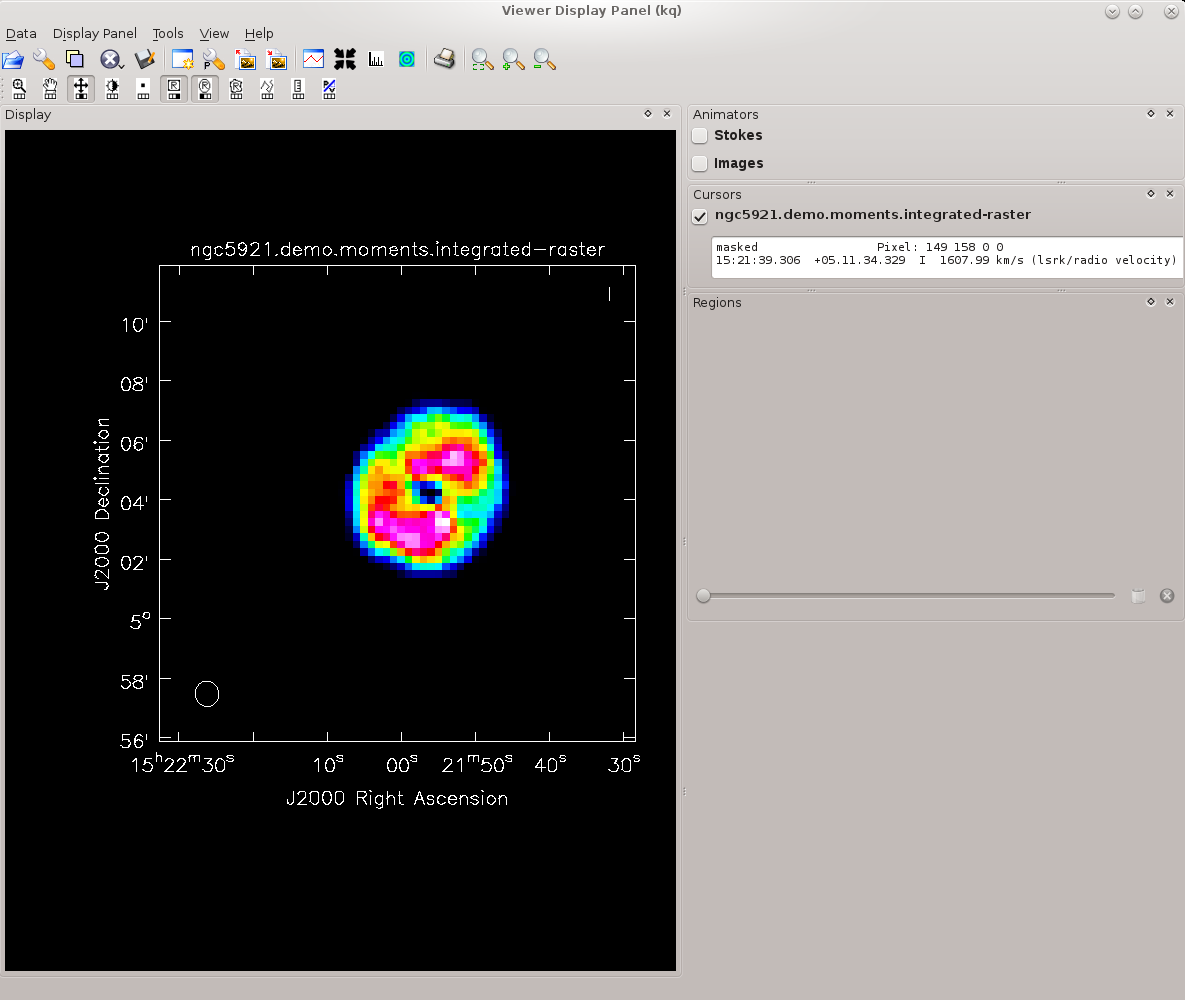
Cube moments are maps of weighted sums along the velocity axis. In CASA, they are generated by the task immoments. The zeroth moment (moments = 0) is a sum of intensities along the velocity axis (the integrated intensity map); the first moment (moment = 1) is the sum of velocities weighted by intensity (the velocity field); the second moment (moment = 2) is a map of the velocity dispersion; see the immoments for additional options.
The following example produces maps of the zeroth and first moments, or the integrated intensity and velocity field. The respective measurement sets are the moment zero image ngc5921.demo.moments.integrated and moment one imagengc5921.demo.moments.weighted_coord.
We will do the zeroth and first moments and mask out noisy pixels using hard global limits. We will also collapse along the spectral (channel) axis and include all planes.
immoments(imagename='ngc5921.demo.tclean.image', moments=[0,1], excludepix=[-100, 0.009], axis='spectral', chans='', outfile='ngc5921.demo.moments')
- moments = [0,1] : Do zeroth and first moments
- excludepix = [-100,0.009] : Mask out noisy pixels using hard global limits
- axis = 'spectral' : Collapse along the spectral (channel) axis
- chans = :Include all planes
To examine the moment images, use viewer; the resulting moment zero image is displayed at right. Note that you may have to play with the color map (Data Display Options button in viewer) in order to replicate the image in this tutorial.
viewer(infile='ngc5921.demo.moments.integrated')
Export the Data
To export the (u, v) data and image cube as FITS files, use exportuvfits and exportfits, respectively.
Here's how to export the continuum-subtracted (u, v) data.
exportuvfits(vis='ngc5921.demo.src.split.ms.contsub', fitsfile='ngc5921.demo.contsub.uvfits', datacolumn='corrected', multisource=True)
And now, the FITS cube.
exportfits(imagename='ngc5921.demo.tclean.image', fitsimage='ngc5921.demo.tclean.fits')
The moment maps (or any CASA images) can be similarly exported using exportfits.
Appendix: Python Notes
os.system
os.system allows you to run shell commands from within python / CASA. For example:
import os
os.system('ls -sF')
will give an OS-level listing of the current directory's contents.
os.environ.get
It's worth having a look at the output of the os.environ.get command to understand the python syntax (alternative: os.getenv). You can think of os.environ as a list of operating system environment variables, and get is a method that extracts information about the requested environment variable, here, CASAPATH. Get returns a string of whitespace separated information, and .split() turns that string into a list. The array index [0] extracts the first element of that list, which contains the path.
To illustrate, here is some example python I/O in CASA.
CASA <12>: print os.environ.get('CASAPATH')
/usr/lib64/casapy/30.0.9709test-001 linux local el5bld64b
CASA <13>: print os.environ.get('CASAPATH').split()
['/usr/lib64/casapy/30.0.9709test-001', 'linux', 'local', 'el5bld64b']
CASA <14>: print os.environ.get('CASAPATH').split()[0]
/usr/lib64/casapy/30.0.9709test-001
Last checked on CASA Version 5.7.2.