Image Continuum CASA 6.1.1: Difference between revisions
| Line 286: | Line 286: | ||
== Imaging the Continuum == | == Imaging the Continuum == | ||
<figure id="Cont_before_clean.png"> | <figure id="Cont_before_clean.png"> | ||
[[File:Cont_before_clean.png|thumb|<caption>During interactive {{clean}}, the GUI will show an initial dirty image. From this GUI, create the {{clean | [[File:Cont_before_clean.png|thumb|<caption>During interactive {{clean}}, the GUI will show an initial dirty image. From this GUI, create the {{clean}} mask.</caption>]] | ||
</figure> | </figure> | ||
<figure id="Cont_before_clean_mask.png"> | <figure id="Cont_before_clean_mask.png"> | ||
Revision as of 19:17, 3 May 2017
This guide should be used after completing Prepare the data for Imaging. You should have created calibrated_final.ms prior to proceeding.
Check CASA version
It is highly recommended that the same CASA version used for calibration is used for imaging. This template is not recommended to be used in a CASA version <4.4. If CASA closes, change the version of CASA you’re using by either downloading the proper version from Obtaining CASA or initializing the suggested version.
# in CASA
import re
if casadef.casa_version < '4.4.0' :
sys.exit("Please use CASA version greater than or equal to 4.4.0 with this script")
Image Parameters
Before imaging, you should set a few key parameters that you will use throughout the rest of the script.
Set the field id for the science target you are interested in imaging. Check the ALMA Observing Tool (OT) as well as the listobs file generated to find this information. If you are imaging a mosaic then you will have to select all the fields as field = ‘3~25’. You can also set the field variable to the name of the source, as long as it matches the name in the listobs file. Do not leave the field parameter blank.
# in CASA
# update for your ms
field='0'
Uncomment the imagermode that is relevant to your dataset. Set the phase center by field id or coordinates if you are imaging a mosaic. Check the spatial setup in the weblog (for pipeline calibrations) or qa2 report (for manual calibrations) to find the phase center. You should choose the central field for the phase center in order to get the best results.
If you are unsure which field to use for the phase center after looking at the weblog then you may use the following Analysis Utilities command:
au.pickCellSize(‘calibrated_final.ms',imsize=True).
This will return a four element array with that contains the calculated cell size, the X axis number of pixels, the Y axis number of pixels, and the field number that is most centered in the mosaic. You may use this as the phase center field id in your script.
Another method of finding the central field number is using the task au.plotmosaic(‘calibrated_final.ms’) within CASA. This method will produce a plot of all fields with their corresponding field numbers plotted on the sky. You can also set the phase center with the coordinates at the center of this plot.
# in CASA
# imagermode='csclean' # uncomment if single field
# imagermode='mosaic' # uncomment if mosaic or if combining one 7m and one 12m pointing.
# phasecenter=3 # uncomment and set to field number for phase
# center. Note lack of ''. Use the weblog to
# determine which pointing to use. Remember that the
# field ids for each pointing will be re-numbered
# after your initial split. You can also specify the
# phase center using coordinates, e.g.,
# phasecenter='J2000 19h30m00 -40d00m00'
<figure id="Calibrated_final_AmpVsUVWave.png">
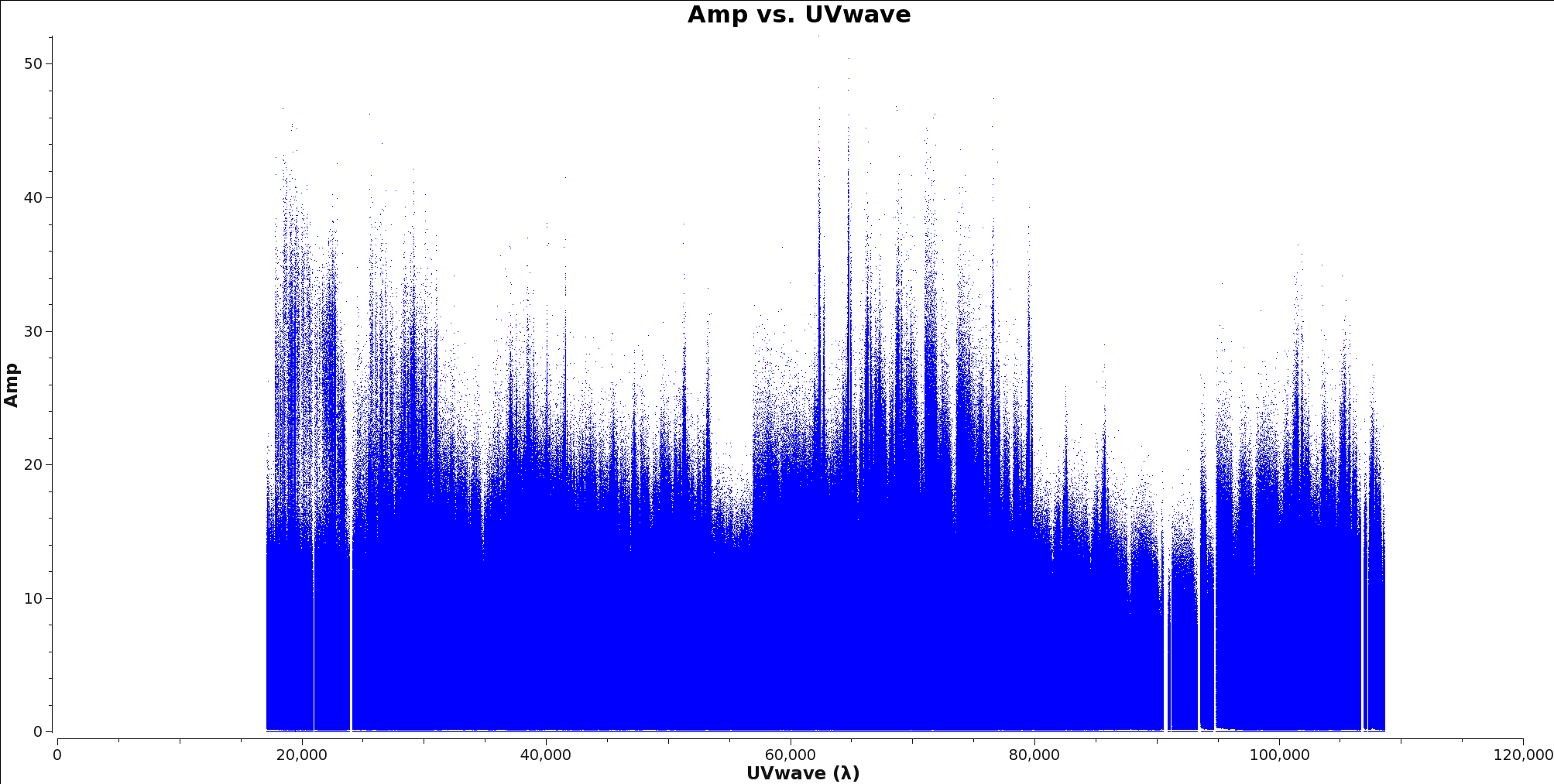
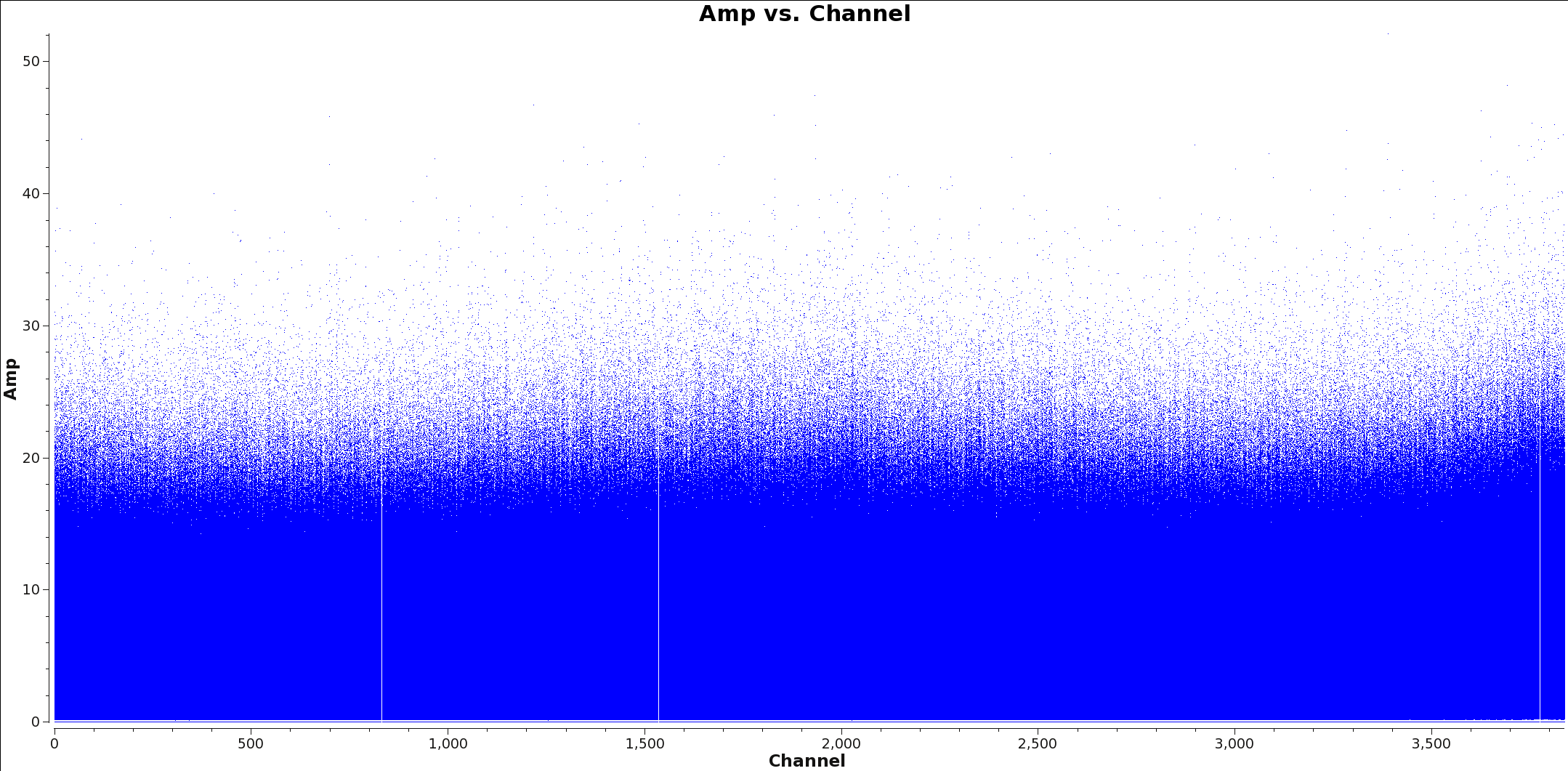
</figure> Next you will determine the cell size. Find the approximate beam size of your observation using the following equation:
206265.0/(longest baseline in wavelengths) ~ Beam Size(arcseconds)
To determine the longest baseline in wavelength use plotms with xaxis=’uvwave’ and yaxis=’amp’ on your ms file. Figure 1 shows an example of this plot. It is recommended you fit 5 to 8 cells per beam when imaging. Therefore you can find the cell size by dividing the Beam Size by at least 5, however it is always better to put more pixels across your beam than too few.
Beam Size/5 ~ Cell Size(arc seconds)
The next step is to determine the image size in pixels. There are two methods of doing this depending on if you are imaging one field or a mosaic.
If this is a single field, the image size can usually be approximated to be the same size as the primary beam of the telescope. The ALMA 12m primary beam in arcsec scales as 6300 / nu[GHz] and the ALMA 7m primary beam in arcsec scales as 10608 / nu[GHz], where nu[GHz] is the sky frequency. However, if there is significant point source and/or extended emission beyond the edges of your initial images, you should increase the imsize to incorporate more emission.
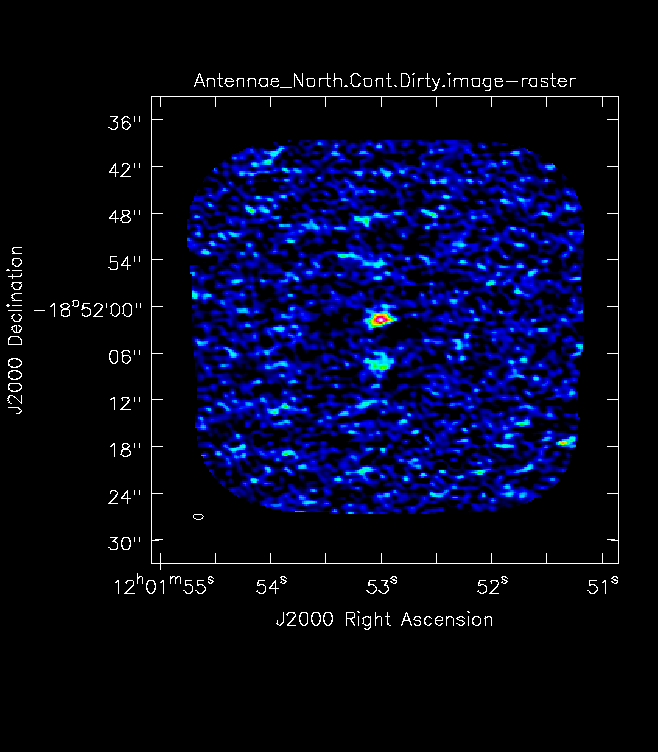
For mosaics, you can get the imsize from the spatial tab of the OT. The parameters "p length" and "q length" specify the dimensions of the mosaic. If you're imaging a mosaic, pad the imsize substantially to avoid artifacts. To know if you have padded enough you should, in your images, be able to see the edges of the outside fields being cut off. <figure id="Antennae_Antennae_North.Cont.Dirty.image.png">
</figure> <figure id="Antennae_North.Cont.Dirty.smallIMSize.image.png">
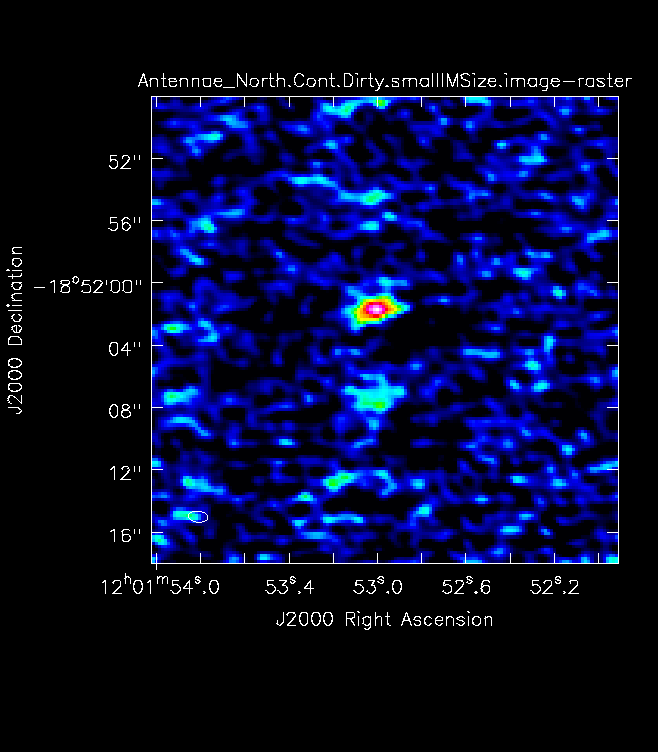
</figure> Note that the imsize parameter is in PIXELS, not arcsec, so you will need to divide the image size in arcsec by the pixel size to determine a value for imsize.
If you would like to check any of these calculations you may use the following command, [au.pickCellSize]('calibrated_final.ms', imsize=True), in CASA. This will return a four element array of the calculated cell size, the x axis imsize, the y axis imsize, and the central field id number.
# in CASA
cell='1arcsec' # cell size for imaging.
imsize = [128,128] # size of image in pixels.
When imaging you will need to set two specific velocity parameters called outframe and veltype. Outframe is the coordinate system used for the observation and can be found in the OT under field setup. A list of acceptable outframes that can be used in CASA can be found at https://help.almascience.org/index.php?/Knowledgebase/Article/View/86/0/what-are-the-frequency-reference-frames-in-casa. Note: heliocentric(hel) is deprecated in CASA. Use barycentric(bary) in this case.
You will also have to set the veltype for the clean command. This variable has only two options available, radio and optical. It is standard to leave this set to ‘radio’ in all projects regardless of the velocity frame used in the project.
# in CASA
outframe='bary' # velocity reference frame. See science goals.
veltype='radio' # velocity type.
The last four parameters that must be set are associated with how clean will weight and clean the data. Weighting determines how much influence data points in the UV plane have on the final image. There are several weighting schemes that can be used such as briggs, natural and uniform weighting.
- Natural: Natural weighting uses the same weights as that of the data weights when collected. This weighting scheme is the default in clean and gives low rms noise to the image but a poor angular resolution.
- Uniform: Uniform weighting will give all points in the UV plane the same influence regardless of their weight. Compared to Natural weighting, it gives more influence to lower weighted cells . Because of this, sidelobes in the image will be reduced but the rms * noise will increase. This type of weighting is best used for point source objects due to the low resolution but at the cost of high rms noise in the image.
- Briggs and the Robust Parameter: This is the most common weighting used and gives a balance between the low rms of natural weighting while also giving the excellent angular resolution of uniform weighting. Briggs weighting will vary the weighting given to UV cells with the robust parameter. Setting this to 2 gives Natural weighting, -2 Uniform weighting, and a number in between a variance of the two. Refer to Brigg's Thesis for more information.
Currently, robust is set to 0.5 which gives a good compromise between natural and uniform weighting. You may find, after imaging, that you have to decrease the rms noise or angular resolution based on the desired performance requested in the proposal. Playing with the robust parameter can affect your final rms noise in the image and also the angular resolution.
The niter and threshold parameters affect when clean will stop the cleaning process. niter is the maximum number of iterations allowed and once this number has reached zero interactive cleaning will finish. Once a certain flux level, set by threshold, has been reached, clean will stop. This is by default set at 0.0mJy but may be increased to prevent cleaning too deeply into the thermal noise.
There are several methods to determine what to set threshold to if you desire to change it. One method is to make a clean mask by setting niter equal to zero, then inspecting the image produced using viewer. Select a box around an area of no emission to determine the rms noise of the image, which should be the approximate value of the threshold parameter. Follow the instructions located in the “Image the Spectral Line Data” section of EVLA Spectral Line Imaging Analysis IRC+10216. If you plan on cleaning non-interactively then you must set threshold to a value other than 0.
# in CASA
weighting = 'briggs'
robust=0.5
niter=1000
threshold = '0.0mJy'
For more information on the various options available in clean, refer to David Wilner’s Imaging and Deconvolution presentation from the 2016 Synthesis Imaging Workshop. Use this table to find specific slides for different parameters you will set below.
| Pixel and Image Size | Slides 40-41 |
| Weighting | Slides 42-48, 60-61 |
| Deconvolution Algorithms | Slides 50-53, 62 |
Create an Averaged Continuum MS
<figure id="Calibrated_final_Field0_Spw0.png">
</figure> <figure id="Calibrated_final_Field0_Spw3.png">
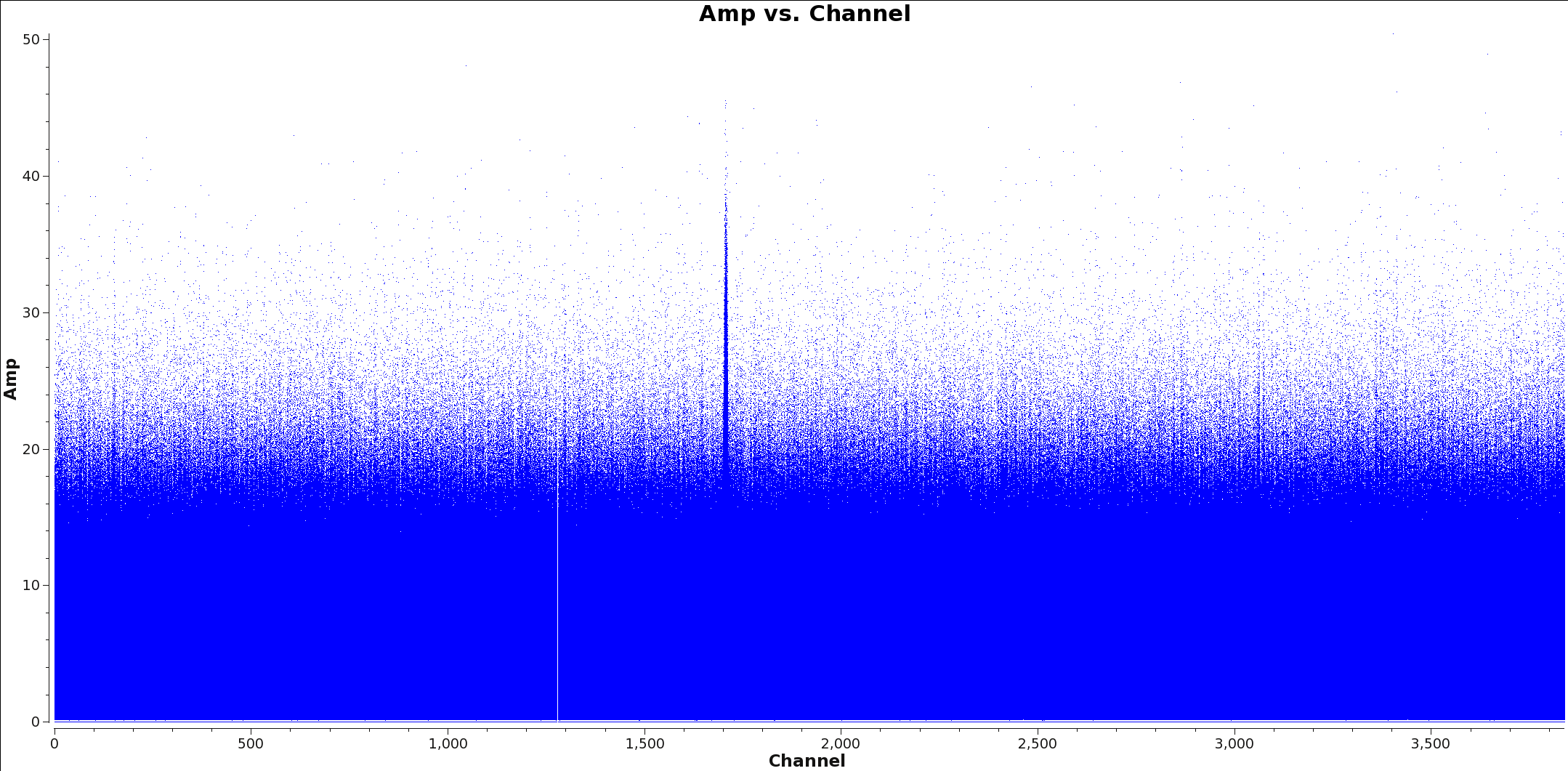
</figure> Create a separate, spectrally-averaged continuum measurement set in order to increase the efficiency of clean/tclean. The process below identifies line free channels and creates this measurement set. The first step is to identify spectral lines in all spectral windows(spw) you will use to make the continuum image. Include all spectral windows with greater than 250 MHz bandwidth in order to maximize the continuum bandwidth. If you find spectral line emission you will need to flag these channels from the spw before imaging.
There are currently two methods to determine which channels have spectral line emission. The first is to simply use CASA to plot Channel vs Amp of each spw and note the channels with emission. You may wish to change the averaging and uvrange to identify extended emission:
# in CASA
plotms(vis=finalvis, xaxis='channel', yaxis='amplitude',
ydatacolumn='data',
avgtime='1e8', avgscan=True, avgchannel='1',
iteraxis='spw' )
The second, and more accurate, method is to use clean to make a quick dirty image cube of channels with niter set to zero and mode=’channel’. Inspecting this channel cube for line emission gives a better defined channel width to flag. The command below gives the basic command to create dirty image cubes. This should be repeated for each spw and science field.
#In CASA
testimagename=’testImage’
field=[‘0’] #list all target fields
spw=[‘0,1,2,3’] #list all target spw’s
for i in field:
for j in spw:
clean(vis=finalvis,
imagename=testimagename+’Field_’+str(i)+’_spw_’+str(j),
field=str(i),
spw=str(j),
# phasecenter=phasecenter, # uncomment if mosaic.
mode='channel',
outframe=outframe,
niter=0,
interactive=True,
cell=cell,
imsize=imsize,
weighting=weighting,
robust=robust,
imagermode=imagermode)
Once you have determined which spws have line emission and which ones you would like to use to form the continuum from your measurements, set the continuum spws to use:
# in CASA
# Set spws to be used to form continuum
contspws = '0,1,2,3'
<figure id="Wt_vs_Freq_spw0123.png">
</figure> The first step to flagging the spectral line channels in your data is to use the flagmanager task to save the state of the data before any flagging is applied. You will need to revert back to the non spectral line flagged dataset when line imaging is done later on. In addition, if you accidentally flag any data you can easily correct this using the *.before_cont_flags file.
# in CASA
flagmanager(vis=finalvis,mode='save',
versionname='before_cont_flags')
Initialize the weights in the ms using initweights.
# in CASA
initweights(vis=finalvis,wtmode='weight',dowtsp=True)
The exact spectral window and channel ranges to flag in the flagchannels variable needs to be specified. For example, if spws 2&3 have a line between channels 1201 and 2199 and spectral windows 0 and 1 are line-free the command would be:
# in CASA
flagchannels='2:1201~2199,3:1201~2199' # modify the channel range for your dataset
<figure id="Calibrated_final_field0_spw0_LineChannelFlagged.png">
</figure> Next, use the task flagdata to apply these flags. After this is done it is good to check that the flags were applied correctly by using plotms to inspect the flagged ms.
# in CASA
flagdata(vis=finalvis,mode='manual',
spw=flagchannels,flagbackup=False)
# check that flags are as expected, NOTE must check reload on plotms
# gui if its still open.
plotms(vis=finalvis,yaxis='amp',xaxis='channel',
avgchannel='1',avgtime='1e8',avgscan=True,iteraxis='spw')
Before imaging the continuum there are several things that can be done to speed up cleaning of the final continuum image. One of these is to spectrally average several channels together within a single continuum window thus reducing the total number of channels while maintaining the same bandwidth.
Refer to [How to Calculate Bandwidth Smearing] for the derivation of the information about bandwidth smearing. Conservative values for various ALMA bands are suggested below.
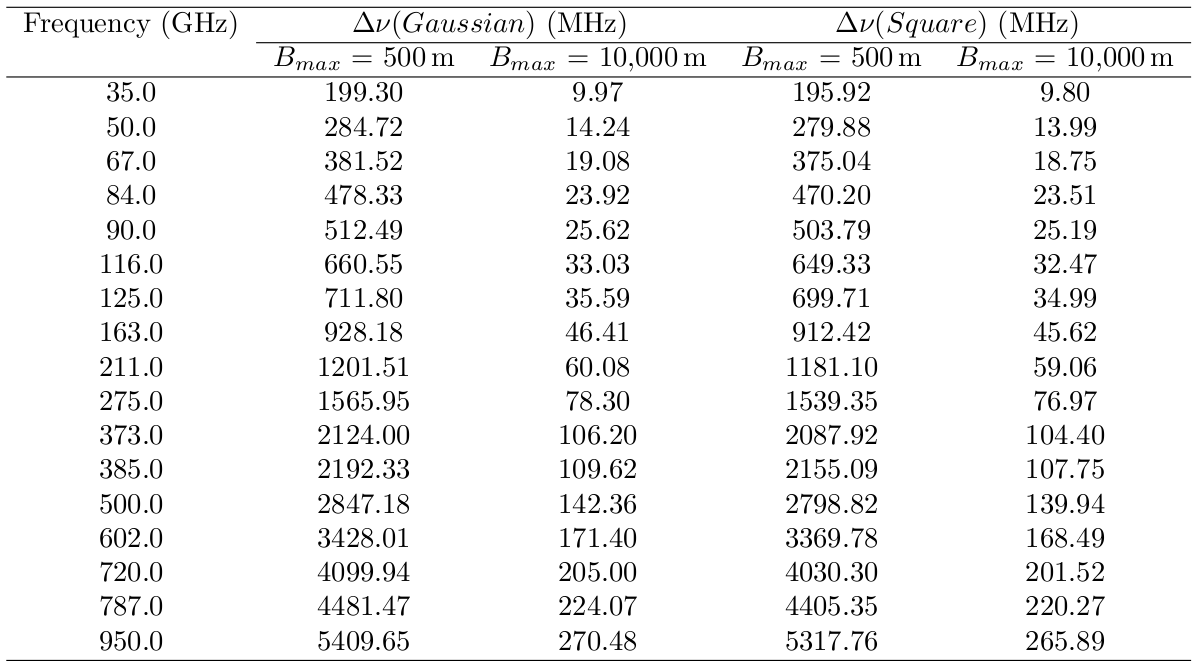
This table lists the maximum bandwidth allowed for a reduction in peak response to a point source over the field of view of 1% for a a square and Gaussian bandpass. This tables lists this for various observing frequencies and baselines for the two bandpass cases. For example, if your observation was at 385 GHz and your maximum baseline was 10,000 meters then for a Gaussian bandpass the maximum bandwidth that can be averaged together is 109.62 MHz bandwidth.
125MHz/ChanWid(MHz) = number of channels to average for bands 3,4,5,6 250MHz/ChanWid(MHz) = number of channels to average for band 7
For example, if you have 128 channels and a channel width of 15.625 MHz in band 6, the way to find the number of channels to average together is
125 MHz / 15.625 MHz = 8 channels
It is always a good check to make sure the number of channels you are averaging together is an integer multiple of the total number of channels original. This will tell you how many channels will remain in the ms. Using the example above averaging every 8 channels of 128 channels leaves 16 final channels each with a channel width of 125MHz.
split averages the number of channels you specify within the width command and will save a new ms file called calibrated_final_cont.ms.
# in CASA
contvis='calibrated_final_cont.ms'
rmtables(contvis)
os.system('rm -rf ' + contvis + '.flagversions')
split2(vis=finalvis,
spw=contspws,
outputvis=contvis,
width=[8,8,8,8], # number of channels to average together. The final channel width should be less than 125MHz in Bands 3, 4, and 6 and 250MHz in Band 7.
datacolumn='data')
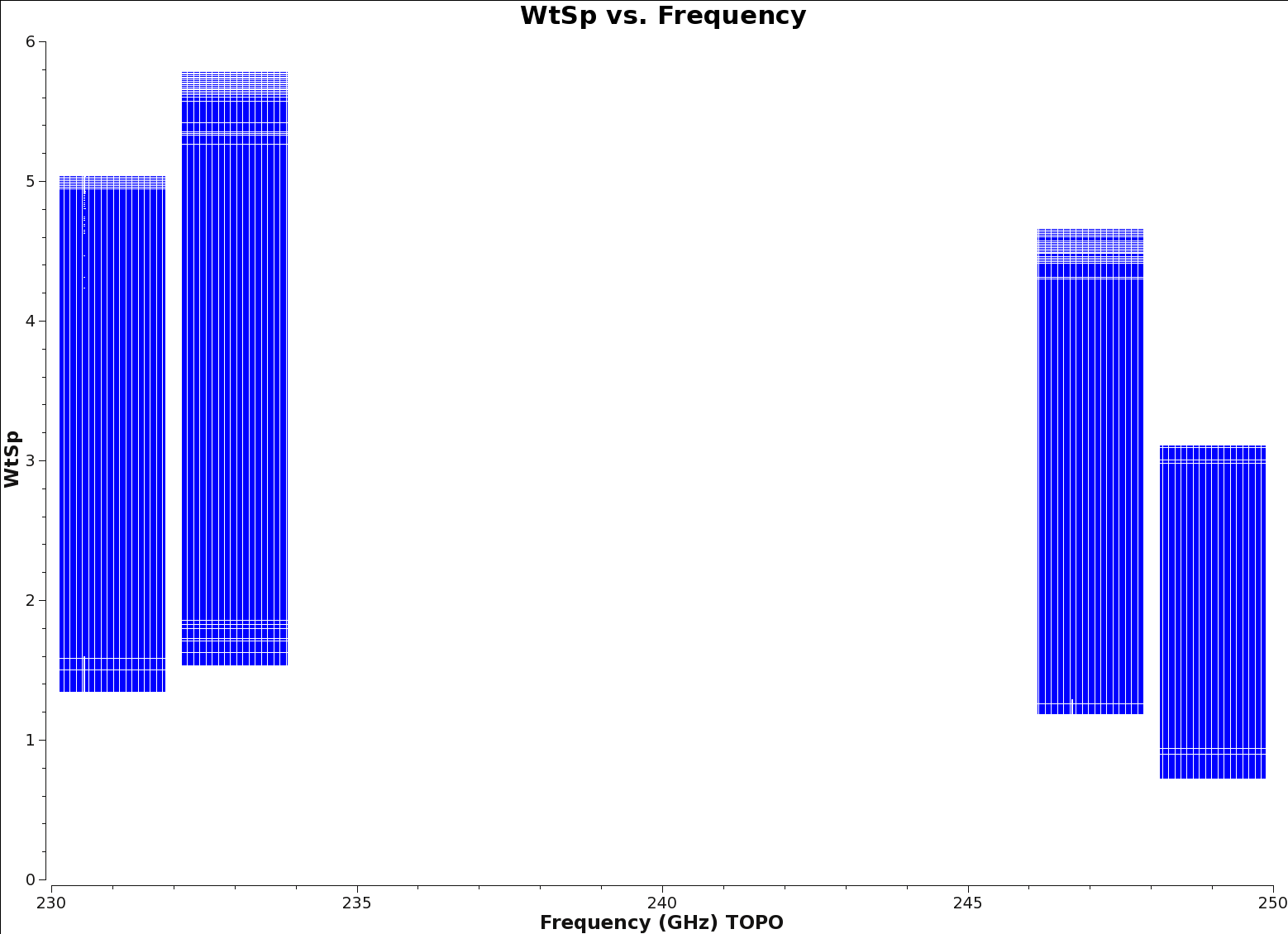
Now you should check the weights of the new continuum measurement set. The ratio of the weights value between Time Domain Mode (TDM) and Frequency Domain Mode (FDM) windows should be approximately equal to the ratio of the channel widths.
# in CASA
# update the antenna and field parameters for your dataset
plotms(vis=contvis, yaxis='wtsp',xaxis='freq',spw='',antenna='DA42',field='0')
Use flagmanager to restore the ms file to its original unflagged state. <figure id="Amp_vs_uvdist.png">
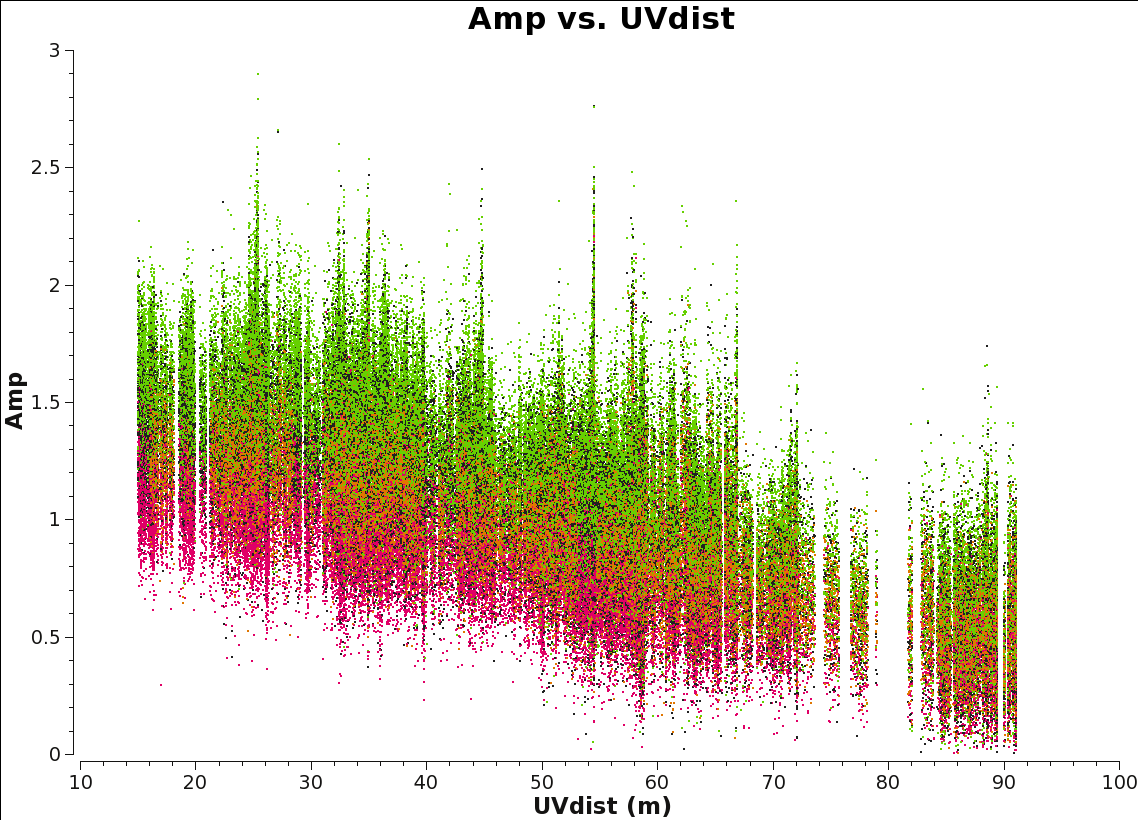
</figure>
# in CASA
# If you flagged any line channels, restore the previous flags
flagmanager(vis=finalvis,mode='restore',
versionname='before_cont_flags')
It is a good practice to inspect the continuum after this in order to confirm it is alright for future cleaning.The uv-coverage should be consistent and you should not see large outliers.
# in CASA
plotms(vis=contvis,xaxis='uvdist',yaxis='amp',coloraxis='spw')
Imaging the Continuum
<figure id="Cont_before_clean.png">
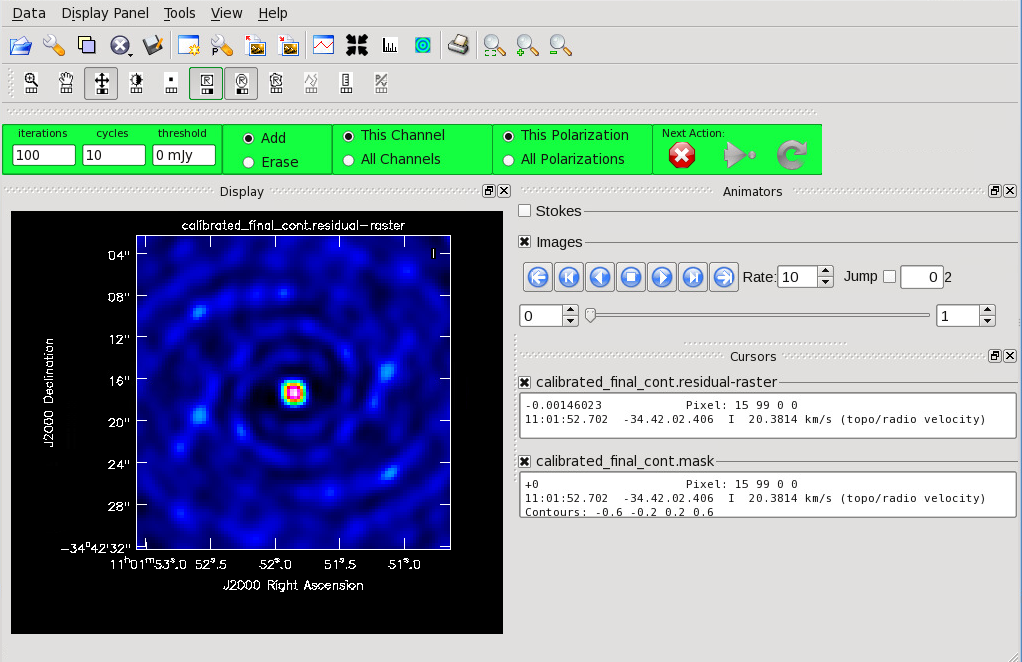
</figure> <figure id="Cont_before_clean_mask.png">
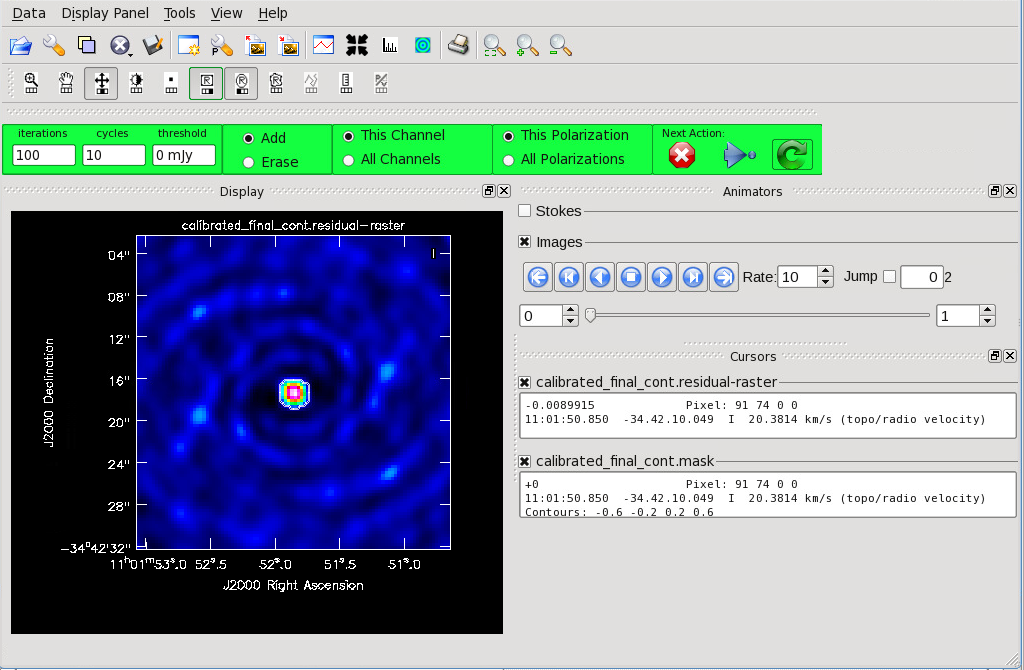
</figure> <figure id="Final_Cont_residual.png">
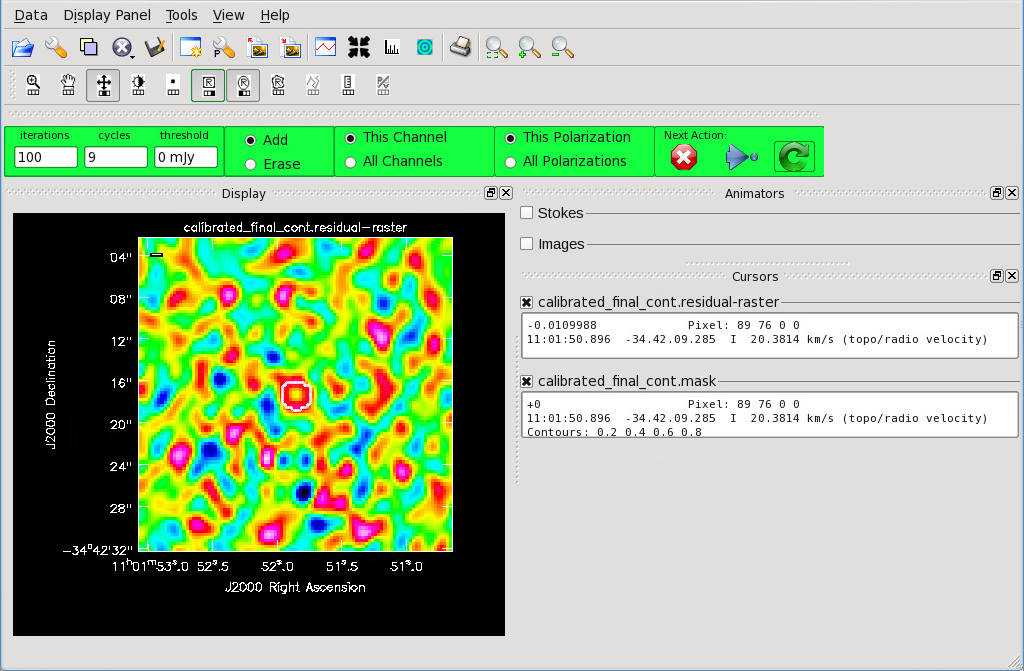
</figure> Now that you have set all of the imaging parameters you will need in clean you can proceed to imaging the continuum. The First Look at Imaging CASAGuide gives an introduction to cleaning and imaging.
# in CASA
contvis = 'calibrated_final_cont.ms'
contimagename = 'calibrated_final_cont'
If you have already run clean on the ms and would like to start over, be sure to use the following commands before cleaning the continuum.
# in CASA
# Clearcal clears the calibrated column of the ms.
# clearcal(contvis)
# Delmod removes any model column put in by clean.
# delmod(contvis)
clean creates a variety of files after it completes. If these files exist in the directory, clean will begin working on the files that already exist. The following command will delete all of these files. You may also change the cont_image_name parameter if you would like to keep the files you created previously.
# in CASA
for ext in ['.flux','.image','.mask','.model','.pbcor','.psf','.residual','.flux.pbcoverage','.pb','.wtsum']:
rmtables(contimagename+ext)
If you are imaging a mosaic, the phasecenter parameter should be set. Refer to the imaging parameters section of this guide to see how to find this for your project.
Type “help clean()” in CASA if you would like to explore the possible parameters of clean. Mode=’mfs’ sets the spectral gridding type to multi-frequency synthesis and creates a continuum image. Leave the psfmode to use during minor cycles at the default ‘clark’ .
clean interactively as the threshold is set at 0 mJy. The mask parameter may also be added if you have an existing file.
# in CASA
clean(vis=contvis,
imagename=contimagename,
field=field,
# phasecenter=phasecenter, # uncomment if mosaic.
mode='mfs',
psfmode='clark',
imsize = imsize,
cell= cell,
weighting = weighting,
robust = robust,
niter = niter,
threshold = threshold,
interactive = True,
imagermode = imagermode)
Figure 11 shows the clean GUI that will appear. If no source is apparent, no cleaning should be done. Press the red “X” to complete the task. If a source is apparent, create a mask around it. Once a mask is created, the green arrow will be illuminated and you can begin the first round of cleaning.
The logger window gives you vital information on the progression of clean. Once the cycle is complete, the residuals will appear in the GUI. You should continue to iterate until the region inside the mask matches the noise outside the mask. You may need multiple cycles depending on the complexity of the source.
The red “X” is the only way clean can be safely exited. Otherwise, your measurement set will likely be corrupted.
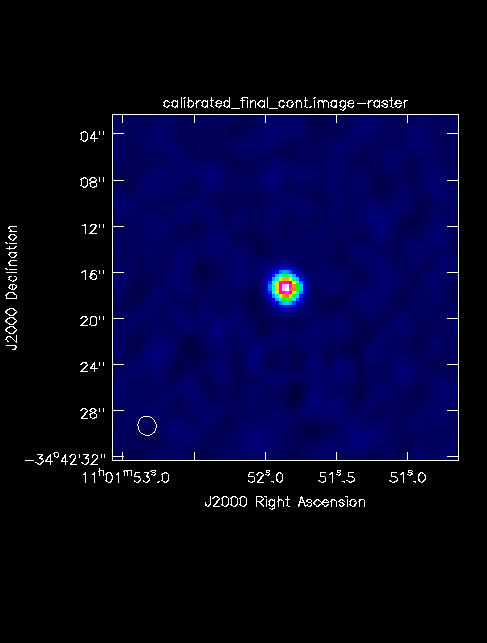
The final continuum image.
After creating the continuum image, check the RMS and beam size to make sure the results match the expected values. Once you are happy with your continuum image(s), you can continue to Self-Calibration Template or Spectral Line Imaging Template. If you do not wish to self-calibrate or create line cubes, continue with this guide to create primary beam corrected images and fits files.
Apply a primary beam correction
<figure id="TW_Hya_Calibrated_final.pbcor.png">
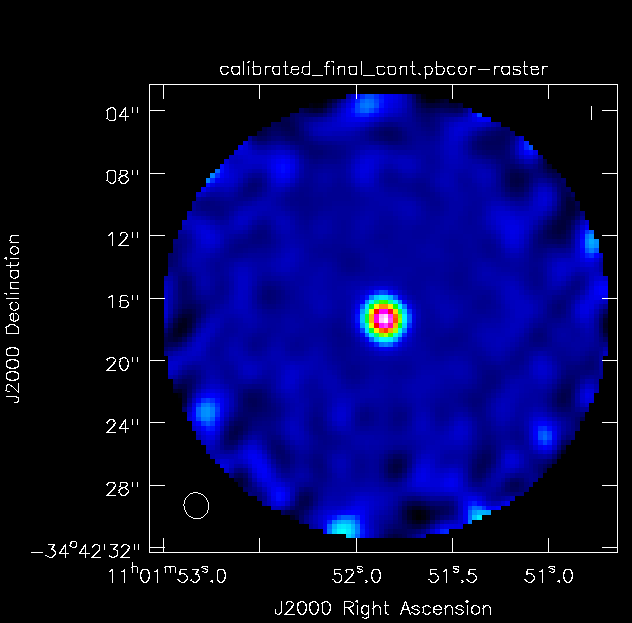
</figure> Once all imaging is finished, diagnostic images will be made. The first of these is a primary beam correction image of the data. Each telescope's primary beam is, for the most part, approximately a Gaussian with small side lobes that can be approximated to zero. Because of this, the center of the beam has much more sensitivity than the edges. To correct for this, flux is added to the edges of the image in order for all pixels across the beam to have the same relative brightness based on the beam pattern.
impbcor is used to produce the primary beam corrected images . You will need the *.flux and *.pbcor files.
# in CASA
import glob
myimages = glob.glob("*.image")
rmtables('*.pbcor')
for image in myimages:
pbimage = image.rsplit('.',1)[0]+'.flux'
outfile = image.rsplit('.',1)[0]+'.pbcor'
impbcor(imagename=image, pbimage=pbimage, outfile = outfile)
Export the images
Use exportfits to create fits files of the *.flux and *.pbcor files.
# in CASA
import glob
myimages = glob.glob("*.pbcor")
for image in myimages:
exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True)
myimages = glob.glob("*.flux")
for image in myimages:
exportfits(imagename=image, fitsimage=image+'.fits',overwrite=True)
Create Diagnostic PNGs
The First Look at Image Analysis guide gives an introduction to a variety of options to begin image analysis, including using immoments to create moment maps. The commands create png files of the continuum image and moment 8 maps.
<source lang="python">
- in CASA
os.system("rm -rf *.png") mycontimages = glob.glob("calibrated*.image") for cimage in mycontimages:
max=imstat(cimage)['max'][0]
min=-0.1*max
outimage = cimage+'.png'
os.system('rm -rf '+outimage)
imview(raster={'file':cimage,'range':[min,max]},out=outimage)