TWHydraBand7 Imaging for CASA 3.3: Difference between revisions
| Line 157: | Line 157: | ||
applycal(vis='TWHydra_cont.ms',field='',gaintable=['self_2.pcal'],calwt=F) | applycal(vis='TWHydra_cont.ms',field='',gaintable=['self_2.pcal'],calwt=F) | ||
</source> | </source> | ||
[[Image:viewer_pcal2.png|thumb|Clean residuals after 2nd phase only self-cal.]] | |||
Now clean again, updating '''mask''' with the most recent mask file. | Now clean again, updating '''mask''' with the most recent mask file. | ||
| Line 175: | Line 177: | ||
Model 0: max, min residuals = 0.00503853, -0.00479908 clean flux 1.47338 | Model 0: max, min residuals = 0.00503853, -0.00479908 clean flux 1.47338 | ||
</pre> | </pre> | ||
It looks like it could use just a little more cleaning, so set '''iterations=50 in the gui'''. | It looks like it could use just a little more cleaning, so set '''iterations=50 in the gui'''. | ||
Revision as of 14:54, 27 May 2011
This tutorial picks up where TWHydraBand7_Calibration leaves off with the fully calibrated split science target ms. If you are starting directly from the TWHYA_Band7_CalibratedData.tar.gz file, untar and unzip it and if necessary rename it TWHydra_corrected.ms.
Prepare Continuum Data for Further Processing
In order to speed up imaging/self-calibration for the continuum data we will average channels together, and then flag the few spectral features. This method works well in the case of only a few spectral lines, while retaining enough channelization to aid in multi-frequency synthesis and being able to remove spectral features in the averaged data. The amount of channel averaging should be adjusted to your particular data. Note for wide bandwidth data it is never a good idea to use the continuum estimate generated by uv-continuum subtraction as your continuum data. This worked OK in the past with the narrow band systems like the VLA and low dynamic range instruments like the SMA, but is not advised for ALMA wide bandwidth, high dynamic range data.
One can also flag the spectral features first but in that case make a backup of the file that is analogous to TWHydra_corrected.ms as you will need it later (unflagged) for spectral line imaging.
# In CASA
split(vis='TWHydra_corrected.ms',outputvis='TWHydra_cont.ms',
spw='0~3:22~3820',width=100,datacolumn='data')
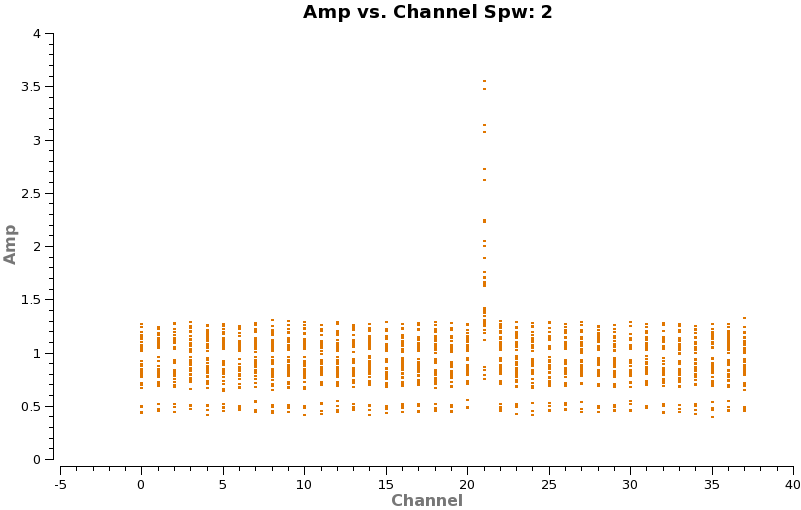
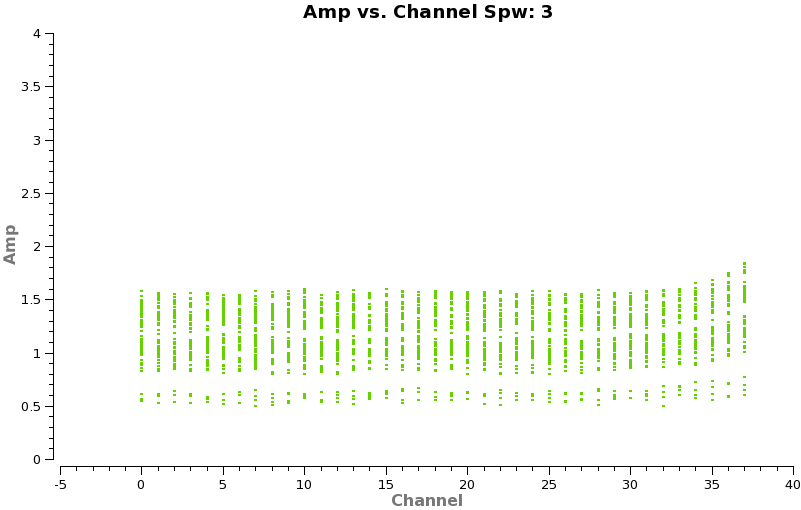
Now make a plot of amplitude vs. channel to see what needs to be flagged
# In CASA
plotms(vis='TWHydra_cont.ms',spw='0~3',xaxis='channel',yaxis='amp',
avgtime='1e8',avgscan=T,coloraxis='spw',iteraxis='spw',xselfscale=T)
The CO(3-2) and HCO+(4-3) lines are obvious. The 4th spw; spw=3 also shows an upswing on the highest channels that is problematic, it was seen on the calibrators too and is likely due to a weak atmospheric feature, we will flag the worst of it.
# In CASA
flagdata(vis='TWHydra_cont.ms',
spw=['0:16~16','2:21~21','3:33~37'])
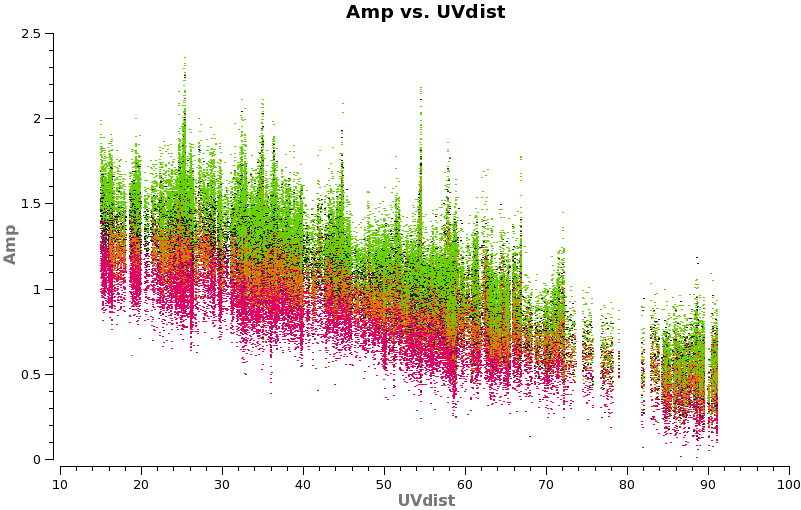
Have a look at the continuum as a function of uv-distance (in the plot below, the 4 spw are shown in different colors on top of each other). If the data are flat as a function of uv-distance the source is completely unresolved. If you see structure (in this case decreasing amplitude with uv-distance) the source is resolved.
# In CASA
plotms(vis='TWHydra_cont.ms',spw='',xaxis='uvdist',yaxis='amp',field='',avgchannel='38',
coloraxis='spw',plotfile='cont_uvplot.png')
Notice that the spw do not have the same amplitude. Unfortunately, the red spw (highest frequency) should be higher than the green one (lowest frequency) because optically thin dust goes as nu^4, and optically thick as nu^2. This inconsistency is due to imperfect amplitude/absolute flux calibration. The self-calibration below will bring them into agreement with each other, but this is the sort of thing that results in considerable absolute flux uncertainty at submillimeter wavelengths. We are working on how to do this better.
Image and Self-Calibrate the Continuum
Initial Clean Image
Make an initial image using clean. We use mode='mfs' to do multifrequency synthesis, in other words grid each uv-channel independently. Since the uv-spacing is a function of frequency, this will actually achieve greater uv-coverage than if all the channels and spws had been averaged in advance. The imsize=100 and cellsize=0.3arcsec were chosen to be appropriate for the Band and antenna configuration. At ALMA Band 7, the Full Width Half Power primary beam is about 20", so we want to make our image at least this big. We've made it a bit larger out to about the 25% power point. For the configuration used to take these data, the resolution is expected to be about 1.5" and it is a good idea to oversample the beam by a factor of 4 or 5 (we used 5 here).
For doing self-calibration it is essential that only "real" signal be included in the image model, thus it is important to make a clean mask, and clean the first iteration rather conservatively (shallowly) so that you are sure that you do not have clean components in your model that do not represent real source emission. In the example below we set interactive=T so you can interactively make the clean mask and stop clean when the signal from the source starts to approach the surrounding "noise" areas in the residual map.
Note: If you start an interactive clean, and then do not make a mask, clean will stop when you tell it to go on because it has nothing to clean. There is no default mask.
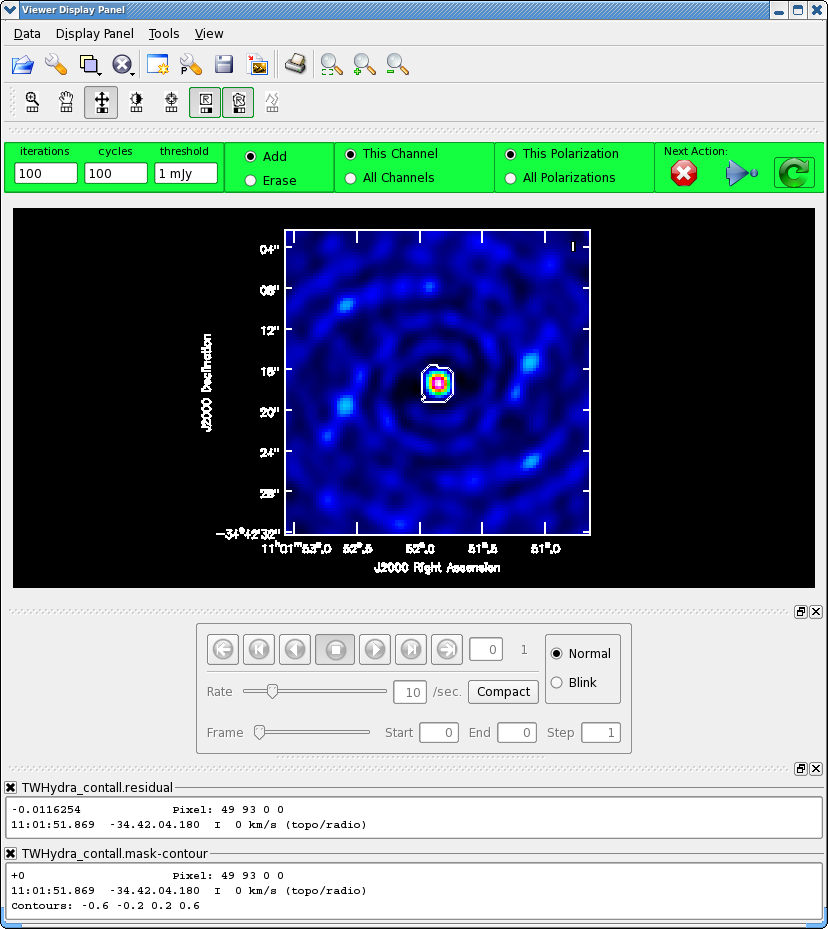
If you are unfamiliar with the CASA viewer it is a good idea to pause here and watch the demo at http://casa.nrao.edu/CasaViewerDemo/casaViewerDemo.html
# In CASA
clean(vis='TWHydra_cont.ms',imagename='TWHydra_contall',
mode='mfs',calready=T,imagermode='csclean',
imsize=100,cell=['0.3arcsec'],spw='',
weighting='briggs',robust=0.5,
mask='',
interactive=T,threshold='1mJy',niter=10000)
Once the Clean Viewer opens, click on the polygon tool with your left mouse button, and draw a polygon around the continuum eimssion with the left mouse button. Double click inside the region with the left mouse button when you are happy with it. It will turn from green to white when the clean mask is accepted. To continue with clean use the "Next action" buttons in the green area on the Viewer Display GUI: The red X will stop clean where you are, the blue arrow will stop the interactive part of clean, but continue to clean non-interactively until reaching the stopping niter or threshold (whichever comes first), and the green circle arrow will clean until it reaches the "iterations" parameter on the left side of the green area. You should chose the green circle arrow. It will clean 100 iterations (number under iteration in the green area on the Viewer gui) and then show the residual image again. The middle mouse button is automatically assigned to the "plus" icon, this adjust the colorscale. If you cannot easily access multiple mouse buttons, you can always assign your one mouse button the icon you want by clicking on it. Adjust the color so you can see it better. Also note that the logger is telling the amount of clean flux recovered so far, the min/max residuals, and the synthesize beam:
Model 0: max, min residuals = 0.0203202, 0.00159066 clean flux 1.31662 Threshhold not reached yet. Beam used in restoration: 1.69902 by 1.59083 (arcsec) at pa 22.3441 (deg)
Already with only 100 iterations, the residuals outside the clean mask are larger than that inside. So we will stop here by hitting the Red X symbol.
Self-calibration
Next we can use the clean model to self-calibrate the uv-data. The process of self-calibration tries to adjust the data to better match the model you give it. That is why it is important to make the model as good as you can. The parameter calready=T ensured that a uv-model of the resulting image was placed in the model data column. This model can then be used to self-calibrate the data using gaincal. We use gaintype='T' to tell it to average the polarizations before deriving solutions which gains us a sqrt(2) in sensitivity. A critical parameter to play with is solint, we use solint=30s to gain a bit of sensitivity. From listobs the integration time of these data are about 10 seconds, using solint=30s then gives us a sqrt(3) in sensitivity. It is often best to start with a larger solint and then work your way down to the integration time if the S/N of the permits. For the same reasons as described in the TWHydraBand7_Calibration tutorial we want to fix up the phases before tackling amplitude by setting calcode='p'.
# In CASA
gaincal(vis='TWHydra_cont.ms',caltable='self_1.pcal',
solint='30s',combine='',gaintype='T',
refant='DV06',spw='',minblperant=4,
calmode='p',minsnr=2)
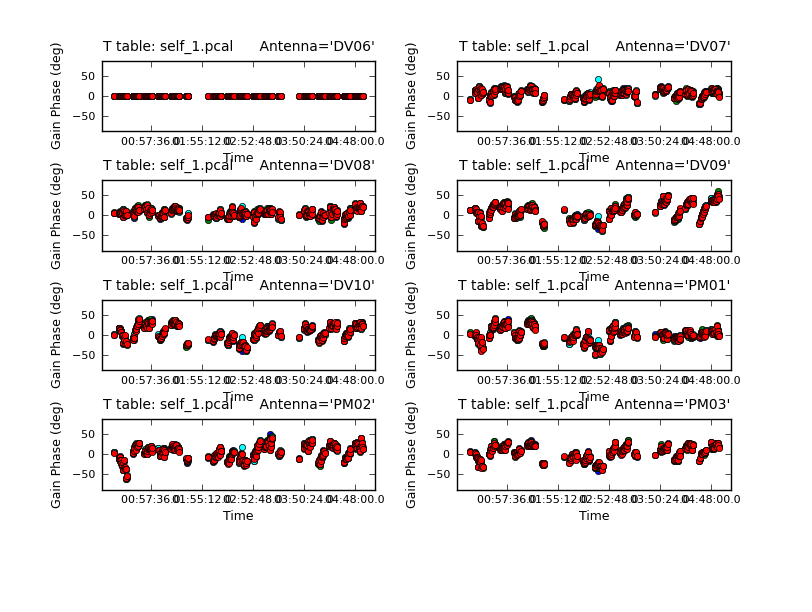
Because combine=, we are getting independent solutions for each spw. If you are desperate for S/N, you can try combining them with combine='spw'. We have plenty of S/N in these data so we keep them separate. Look at the solutions.
# In CASA
plotcal(caltable='self_1.pcal',xaxis='time',yaxis='phase',
spw='',field='',antenna='1~8',iteration='antenna',
subplot=421,plotrange=[0,0,-80,80],figfile='self_1_phase.png')
The magnitude of corrections gives you a sense of how accurate the phase transfer from the calibrators was. You are checking to see that the phases are being well tracked by the solutions, i.e. smoothly varying functions of time within a given scan. Also the solutions for each spw are lying basically on top of each other which indicates that the spw to spw phase offsets are well calibrated already. Overall, these solutions look quite good. If they didn't you could try increasing the solint and/or combining the spw into a single solution.
After checking that you are happy with the solutions, apply them with applycal
# In CASA
applycal(vis='TWHydra_cont.ms',field='',gaintable=['self_1.pcal'],calwt=F)
Next clean the source again, to save time we can start with the clean mask from the last iteration. If you make adjust the clean mask they will be written to the new mask TWHydra_contall_1pcal.mask, and not the old one.
# In CASA
clean(vis='TWHydra_cont.ms',imagename='TWHydra_contall_1pcal',
mode='mfs',calready=T,imagermode='csclean',
imsize=100,cell=['0.3arcsec'],spw='',
weighting='briggs',robust=0.5,
mask='TWHydra_contall.mask',
interactive=T,threshold='1mJy',niter=10000)
Now after the first 100 iterations you see that the residuals inside the clean mask are higher than those outside. This is because flux that had been spread out in the map due to poorly correlated phases has been increased where it belongs on the source. You may also notice that your original clean mask is a little too small. If so, you can make it a bit larger, just by drawing again around where you want the new mask to be. When you double click it will incorporate the new mask (if your box goes awry you can use the delete toggle to erase and redo mask). You might want also want to use the magnifying glass to zoom in. Again you assign mouse buttons but clicking with it on the icon you want. Clean another 100 iterations and stop (for total of 200).
Now open initial and 1st self-cal images to compare them.
# In CASA
Now that we have a better model, we solve for solutions again. This time we will try making the solint smaller -- the integration time is about 10 seconds, so we set solint='int' to have it use the integration time.
# In CASA
gaincal(vis='TWHydra_cont.ms',caltable='self_2.pcal',
solint='int',combine='',gaintype='T',
refant='DV06',spw='',minblperant=4,
calmode='p',minsnr=2)
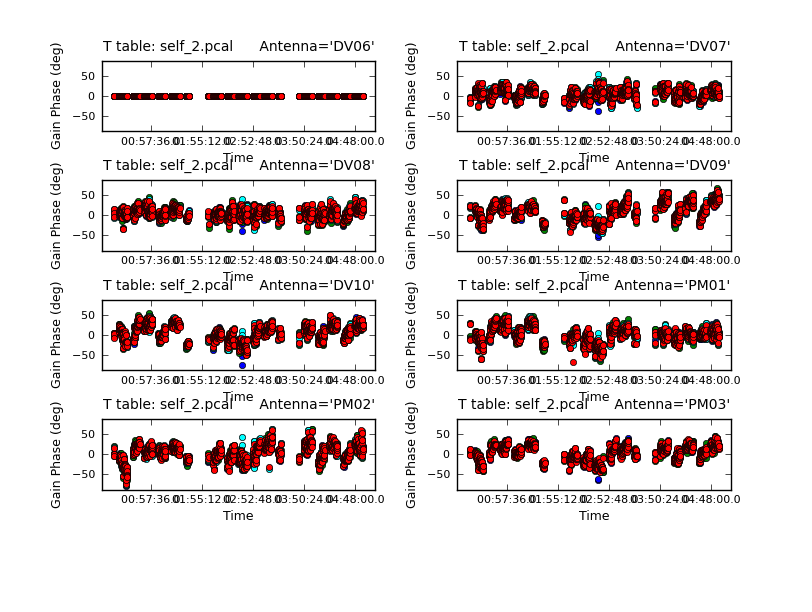
Now plot the new solutions.
# In CASA
plotcal(caltable='self_2.pcal',xaxis='time',yaxis='phase',
spw='',field='',antenna='1~8',iteration='antenna',
subplot=421,plotrange=[0,0,-80,80],figfile='self_2_phase.png')
The phases on this shorter solint still seem to be tracking well so we will apply it and make a new image and check to see if there is improvement.
# In CASA
applycal(vis='TWHydra_cont.ms',field='',gaintable=['self_2.pcal'],calwt=F)
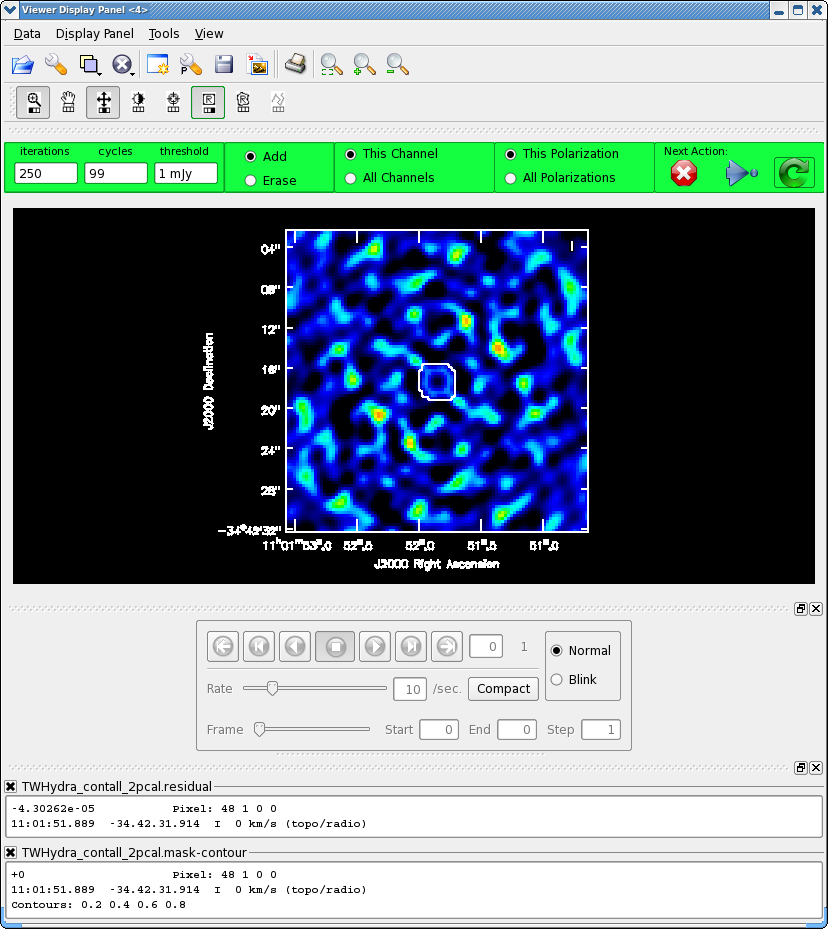
Now clean again, updating mask with the most recent mask file.
# In CASA
clean(vis='TWHydra_cont.ms',imagename='TWHydra_contall_2pcal',
mode='mfs',calready=T,imagermode='csclean',
imsize=100,cell=['0.3arcsec'],spw='',
weighting='briggs',robust=0.5,
mask='TWHydra_contall_1pcal.mask',
interactive=T,threshold='1mJy',niter=10000)
After 200 iterations
Model 0: max, min residuals = 0.00503853, -0.00479908 clean flux 1.47338
It looks like it could use just a little more cleaning, so set iterations=50 in the gui.
Clean used 50 iterations to approach a threshhold of 0.00166574 0.0115307 Jy <- cleaned in this cycle for model 0 Final maximum residual = 0.00328126 Model 0: max, min residuals = 0.00328126, -0.00321554 clean flux 1.48492
The small number cleaned this cycle indicates that clean is converging and its about time to stop. Also the residuals in the clean mask are now smaller than the residuals outside (i.e. the noise) also indicating its time to stop by hitting the Red X on the gui.
# In CASA
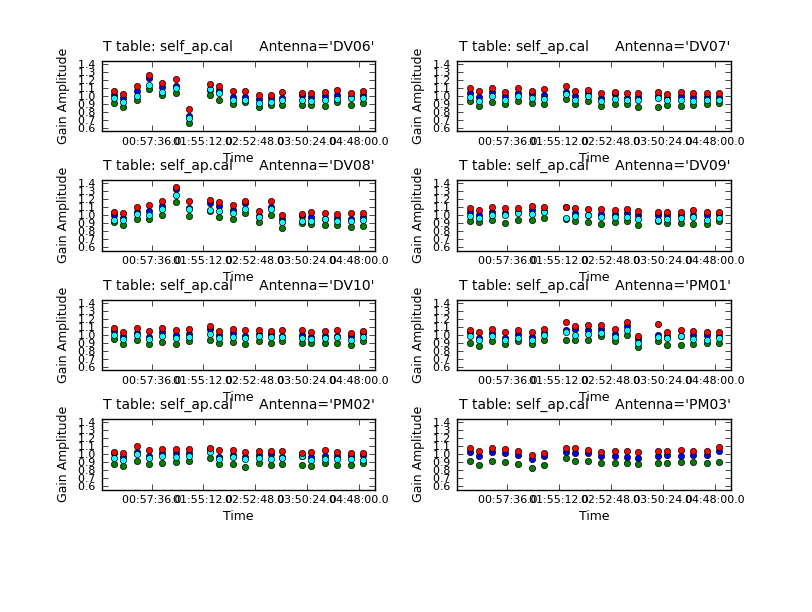
This is only a small improvement suggesting we have gone as far as we can with phase self-cal. Now we will apply the best phase solution table on-the-fly while solving for the amplitude self-cal solutions. Since amplitude changes more slowly and is less constrained than phase, we use a longer solint='inf', as long as combine= this means to do one solution per scan.
# In CASA
gaincal(vis='TWHydra_cont.ms',caltable='self_ap.cal',
solint='inf',combine='',gaintype='T',
refant='DV06',spw='',minblperant=4,
gaintable=['self_2.pcal'],
calmode='ap',minsnr=2,)
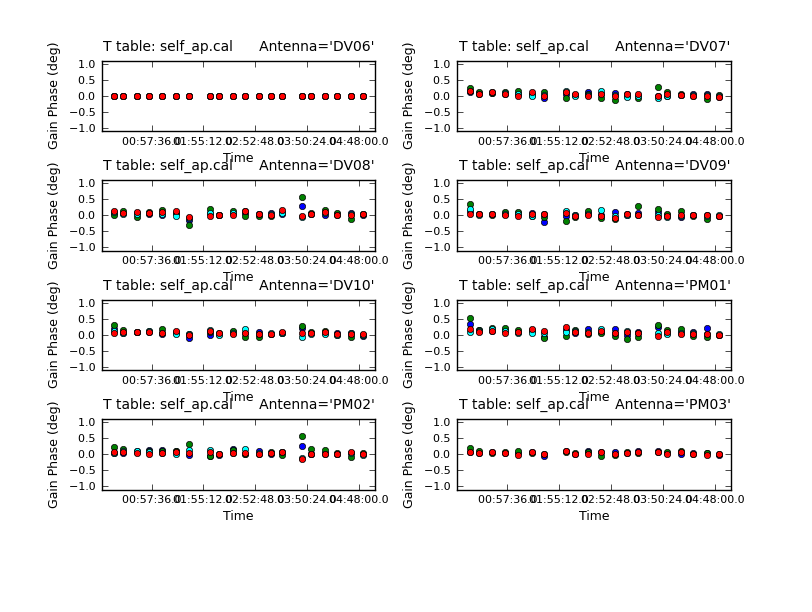
Again its useful to look at the residual phase in the amplitude calibration table.
# In CASA
plotcal(caltable='self_ap.cal',xaxis='time',yaxis='phase',
spw='',field='',antenna='1~8',iteration='antenna',
subplot=421,plotrange=[0,0,-1,1],figfile='self_ap_phase.png')
The residuals are very small as expected. Now look at the amplitude solutions.
# In CASA
plotcal(caltable='self_ap.cal',xaxis='time',yaxis='amp',
spw='',field='',antenna='1~8',iteration='antenna',
subplot=421,plotrange=[0,0,0.6,1.4],figfile='self_ap_amp.png')
For the most part these look very good except one rather large deviation on DV06. We will need to look at this time in the calibrated data. Apply both the final phase and the amplitude calibration tables.
# In CASA
applycal(vis='TWHydra_cont.ms',field='',gaintable=['self_2.pcal','self_ap.cal'],calwt=F)
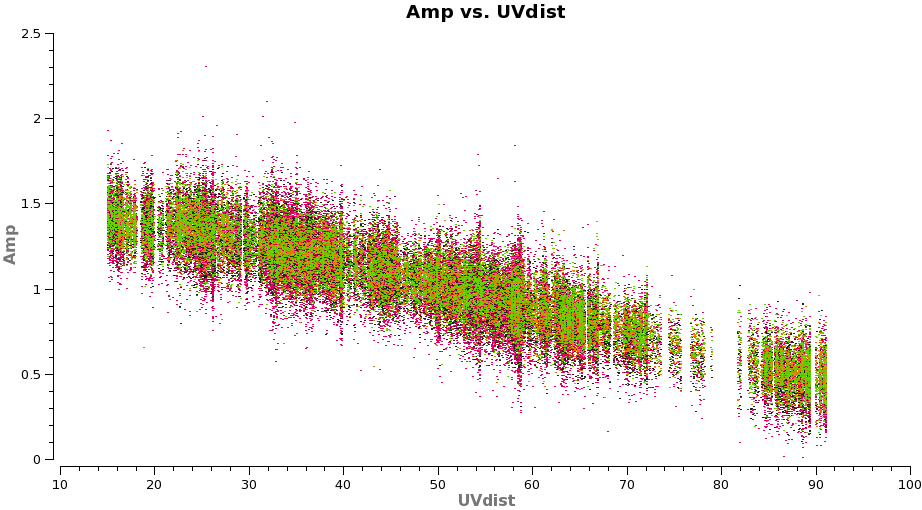
Plot the corrected data as a function of uv-distance. It is always a good idea to check this after an amplitude self-calbration
# In CASA
plotms(vis='TWHydra_cont.ms',xaxis='uvdist',yaxis='amp',
avgchannel='38',ydatacolumn='corrected',coloraxis='spw',
plotfile='selfcal_uvdist.png')
You will see that this looks much better than before.
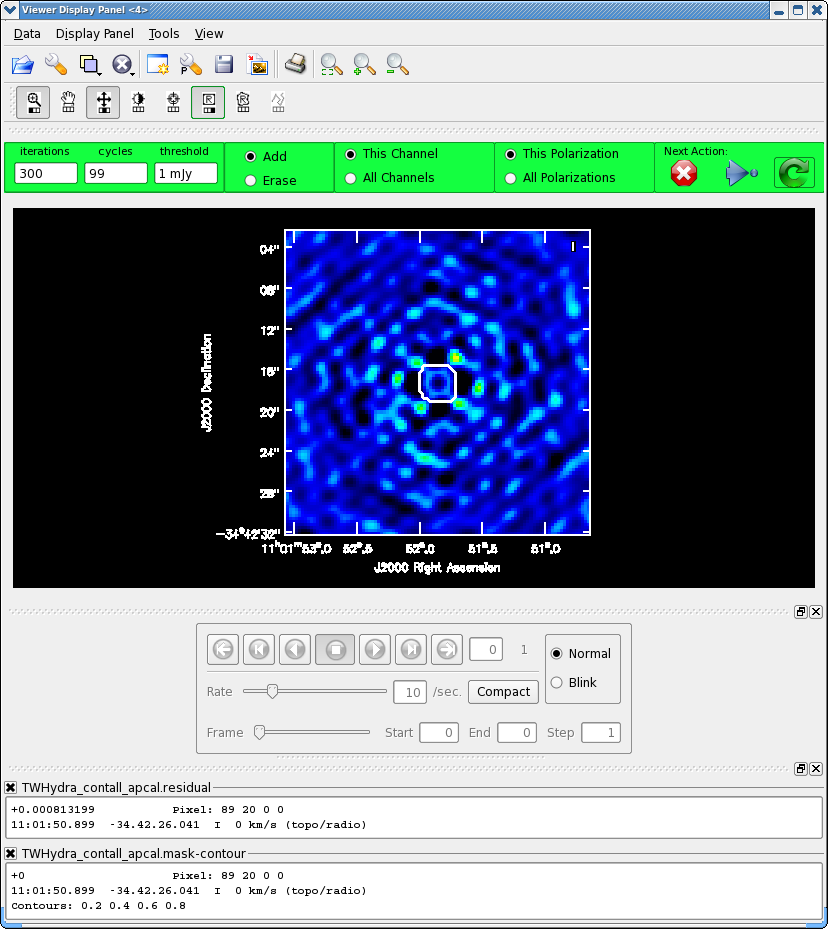
Now make the final continuum image.
# In CASA
clean(vis='TWHydra_cont.ms',imagename='TWHydra_contall_apcal',
mode='mfs',calready=T,imagermode='csclean',
imsize=100,cell=['0.3arcsec'],spw='',
weighting='briggs',robust=0.5,
mask='TWHydra_contall_2pcal.mask',
interactive=T,threshold='1mJy',niter=10000)
We know we need to clean at least 200 iterations, so set this in the gui and hit green arrow. After this notice how much better the residual looks. Clean another 100 iterations. The "circular pattern", and 4 bright spots at each corner you see now in the residual is likely due to non-closing amplitude errors that cannot be removed with an antenna based self-calibration, so this is as good as it gets with pure self-cal. Hit the Red X to stop clean.
Clean used 300 iterations to approach a threshhold of 0.00105507 0.0061808 Jy <- cleaned in this cycle for model 0 Final maximum residual = 0.00242926 Model 0: max, min residuals = 0.00238278, -0.00242926 clean flux 1.48201
Apply self-calibration to Full Dataset and Split Line Data
Now apply the self-calibration derived from the continuum emission to the full unchannel-averaged data.
# In CASA
applycal(vis='TWHydra_corrected.ms',gaintable=['self_2.pcal','self_ap.cal'],calwt=F)
Split off the CO(3-2) spectral line data in spw=2 and the HCO+(4-3) data in spw=0 for further processing.
# In CASA
split(vis='TWHydra_corrected.ms',outputvis='TWHydra_CO3_2.ms',datacolumn='corrected',spw='2')
# In CASA
split(vis='TWHydra_corrected.ms',outputvis='TWHydra_HCOplus.ms',datacolumn='corrected',spw='0')
Continuum Subtraction
Next we need to subtract the continuum emission from the spectral line data. It is best to do this in the uv-plane. The first step is to make a channel plot of the spectral data to get the line-free channel ranges for both spectral lines. Since the line emission is simple (one spectral line per spw) and there are more than enough line-free channels we do not need to be very precise just stay well away from the line and whenever possible it is best to chose line free-channel ranges on either side of the line for the best possible baseline subtraction.
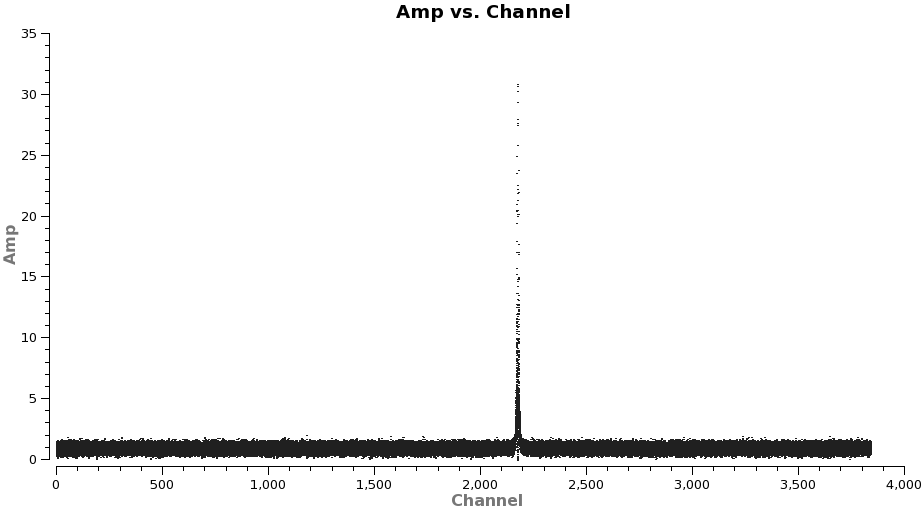
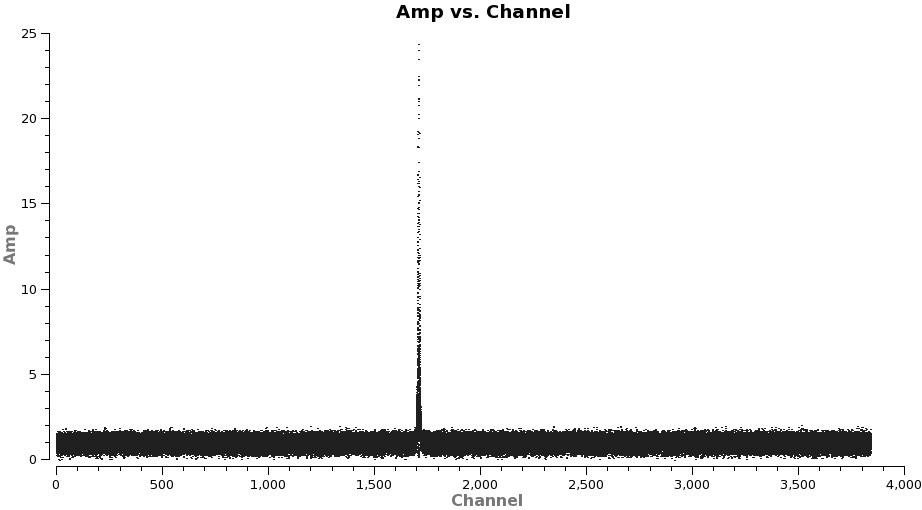
Note: after the split above, the original spw ids will be relabeled as 0 in each file because there is only one spectral window in each file.
# In CASA
plotms(vis='TWHydra_CO3_2.ms',spw='0',xaxis='channel',yaxis='amp',
avgtime='1e8',avgscan=T,coloraxis='spw',plotfile='CO3_2_channel.png')
It is a good idea to stay a little away from the very edges of the baseband which can be noisy. The channel ranges 20~2000 and 2400~3800 look like good choices for CO(3-2).
# In CASA
plotms(vis='TWHydra_HCOplus.ms',spw='0',xaxis='channel',yaxis='amp',
avgtime='1e8',avgscan=T,coloraxis='spw',plotfile='HCOp4_3_channel.png')
The channel ranges 20~1500 and 1900~3800 look like good choices for HCO+(4-3).
Next use the identified line-free channel ranges and uvcontsub2 to subtract the continuum model from the line emission.
# In CASA
uvcontsub2(vis='TWHydra_CO3_2.ms',fitorder=1,fitspw='0:20~2000,0:2400~3800')
# In CASA
uvcontsub2(vis='TWHydra_HCOplus.ms',fitorder=1,fitspw='0:20~1500,0:1900~3800')
Spectral Line Imaging
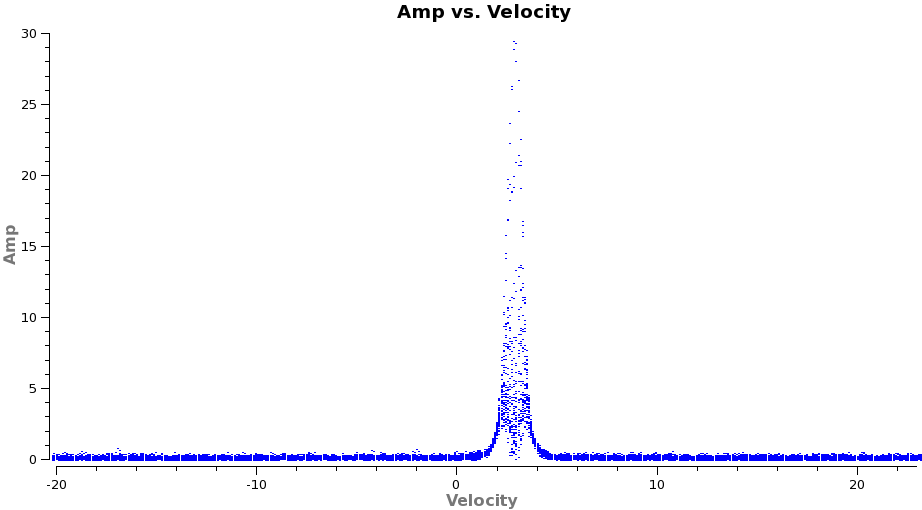
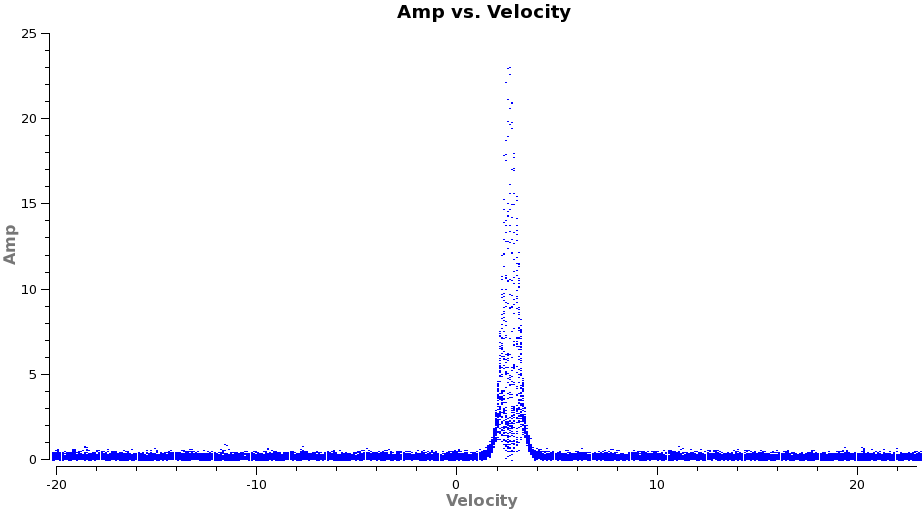
Now we are ready to make cubes of the line emission. The imaging parameters are similar to the continuum except for those dealing with the spectral setup: mode, start, width, nchan, restfreq, and outframe parameters. When making spectral images you have three choices for the mode parameter: channel, velocity, and frequency. Data are taken using constant frequency channels. For spectral line analysis its often more useful to have constant velocity channels, and this is also the best way to make images of multiple lines with the exact same channelization for later comparison. For mode=velocity', the desired start and width also need to be given in velocity units for the desired output frame.
It is important to note that ALMA does not do on-line Doppler Tracking and the native frame of the data is TOPO. If you do not specify outframe the output cube will also be in TOPO, which is not very useful for spectral line work. The Doppler Shift is taken out during the regridding to the desired outframe in clean or alternatively it can be done separately by the cvel task.
To see what velocity parameters you want to set in clean it is useful to make a plot in plotms in the desired output frame. To make it easier to see, we set a plotrange since we know the LSRK velocity of TW Hya is about 2.88 km/s. Note these plots take a little longer because of the frame shift.
# In CASA
plotms(vis='TWHydra_CO3_2.ms.contsub',xaxis='velocity',yaxis='amp',
avgtime='1e8',avgscan=T,transform=T,freqframe='LSRK',
restfreq='345.79599GHz',plotrange=[-20,23,0,0],plotfile='CO3_2_vel.png')
# In CASA
plotms(vis='TWHydra_HCOplus.ms.contsub',xaxis='velocity',yaxis='amp',
avgtime='1e8',avgscan=T,transform=T,freqframe='LSRK',
restfreq='356.7342GHz',plotrange=[-20,23,0,0],plotfile='HCOp4_3_vel.png')
From these plots it looks like imaging the cubes from about -5 to +8 km/s will encompass the line, but still provide several line-free channels on either side so the noise level can be estimated. The channel width is 122 kHz, which at 345.79599 GHz is 0.106 km/s. Recall that the spectral resolution is a factor of two poorer. We will use a velocity channel width of 0.12 km/s for a little oversampling, but the spectral resolution remains 0.2 km/s.
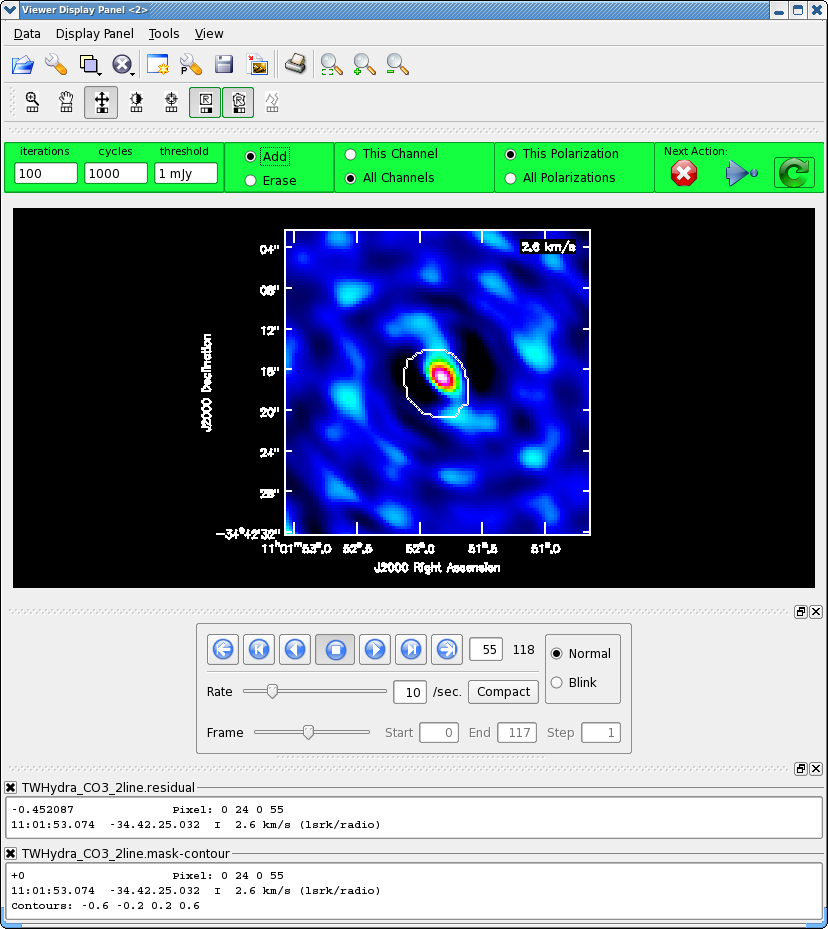
As before, it is very important that you make a clean mask. There are many ways to do this ranging from the complicated to simple. For this example we will make a single clean mask that encompasses the line emission in every channel and apply it to all channels. This is much better than no clean mask, though not quite as good as making an individual mask for each channel. For some sources, one can use the continuum mask to clean the line, however TW Hya is a bit curious in that the continuum emission is not as extended as the line emission (Hughes et al. 2008).
# In CASA
clean(vis='TWHydra_CO3_2.ms.contsub',imagename='TWHydra_CO3_2line',
imagermode='csclean',calready=T,spw='',
imsize=100,cell=['0.3arcsec'],
mode='velocity',start='-4km/s',width='0.12km/s',nchan=118,
restfreq='345.79599GHz',outframe='LSRK',
weighting='briggs',robust=0.5,
mask='',
interactive=T,threshold='1mJy',niter=100000)
It will take a little while to grid the cube, when the interactive viewer opens, use the tapedeck to cycle through the channels. You should see a progress of line emission in channels 40 to 70 or so.
- Go to channel 57 (about line center)
- Select the polygon tool with the left mouse button
- Select the "All Channels" toggle in the green area
- Draw a polygon around the channel 57 emission, double click inside to activate.
- Now go to channel 60, this is the channel with most extent to the SE, draw another polygon that encompasses this emission if necessary,double click inside to activate.
- Now go to channel 55, this is the channel with most extent to the NW, draw another polygon that encompasses this emission if necessary,double click inside to activate.
The figure shows what the final clean box should look like. Now clean using the green circle arrow. You can watch the progress in the logger. When 100 iterations are done, the viewer will show the residual map for each channel. Cycle through the channels and see whether you are still happy with the clean box in each signal channel. The "erase button" can help you fix mistakes. If necessary adjust. It is often useful to adjust the colorscale with the "plus" symbol icon. Keep going until the residual emission looks like the surrounding noise. If it seems to be going slowly, increase the iterations to 300 for a round or two. I stopped cleaning (red x) when the threshold was 0.063 Jy/beam.
Now repeat for the HCO+(4-3) line.
# In CASA
clean(vis='TWHydra_HCOplus.ms.contsub',imagename='TWHydra_HCOplusline',
imagermode='csclean',calready=T,spw='',
imsize=100,cell=['0.3arcsec'],
mode='velocity',start='-4km/s',nchan=118,width='0.12km/s',
restfreq='356.7342GHz',outframe='LSRK',
weighting='briggs',robust=0.5,
mask='',
interactive=T,threshold='1mJy',niter=100000)
Image Analysis
Moment Maps
Next we will make moment maps for the image cubes.
To determine channel ranges it is useful to open cube in viewer as a raster. Using the statistics tool the rms in a line free channel is about 25 mJy/beam. Now open the cube again but as a contour map and set the Unit Contour Level to 0.025 (this is in Jy/beam) and in Relative Contour levels set [3]. i.e. this is showing 3sigma contours. Now find out where the CO(3-2) emission drops below the 3sigma level.
For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity.
# In CASA
immoments(imagename='TWHydra_CO3_2line.image',moments=[0],
outfile='TWHydra_CO3_2line.image.mom0',
chans='40~76')
For higher order moments it is very important to set a conservative flux threshold. Typically something like 10sigma works well.
# In CASA
immoments(imagename='TWHydra_CO3_2line.image',moments=[1,2],
outfile='TWHydra_CO3_2line.image.mom',
chans='40~76',includepix=[0.25,100])
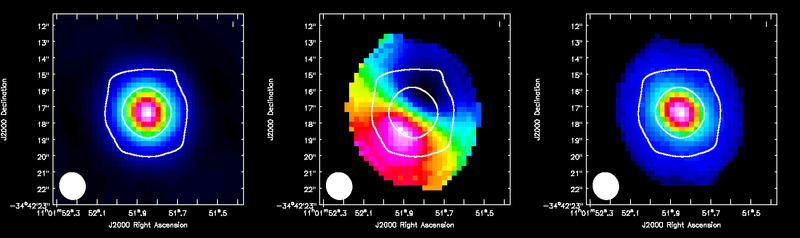
ALMA CO(3-2) moment maps, with white continuum contours at 3 and 100 sigma. From left to right: integrated intensity, intensity weighted velocity field, intensity weighted velocity dispersion are shown.
Repeat for HCO+(4-3) which has a bit narrower velocity extent than the CO(3-2), but similar rms noise.
# In CASA
immoments(imagename='TWHydra_CO3_2line.image',moments=[0],
outfile='TWHydra_CO3_2line.image.mom0',
chans='43~74')
# In CASA
immoments(imagename='TWHydra_CO3_2line.image',moments=[1,2],
outfile='TWHydra_CO3_2line.image.mom',
chans='43~74',includepix=[0.25,100])
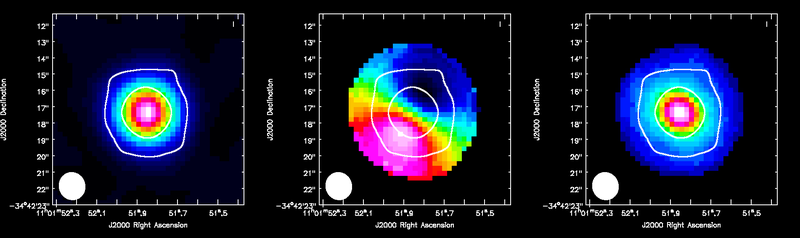
ALMA HCO+(4-3) moment maps, with white continuum contours at 3 and 100 sigma. From left to right: integrated intensity, intensity weighted velocity field, intensity weighted velocity dispersion are shown.
Channel Maps
Using the Viewer you can make channel map figures. Start the Viewer and then open the CO(3-2) cube as a raster image and then the continuum as a contour image. Then we use the "wrench" icon and "P wrench" icons to set up the channel images, contour levels etc.
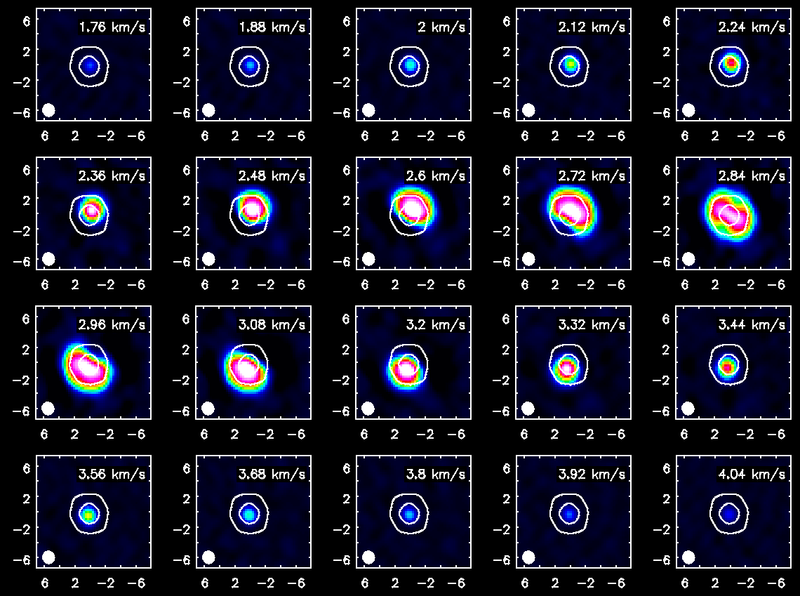
Channel maps of the CO(3-2) emission with white continuum contours at 3 and 100 sigma.
The setup windows looked like this
Repeat for HCO+(4-3)
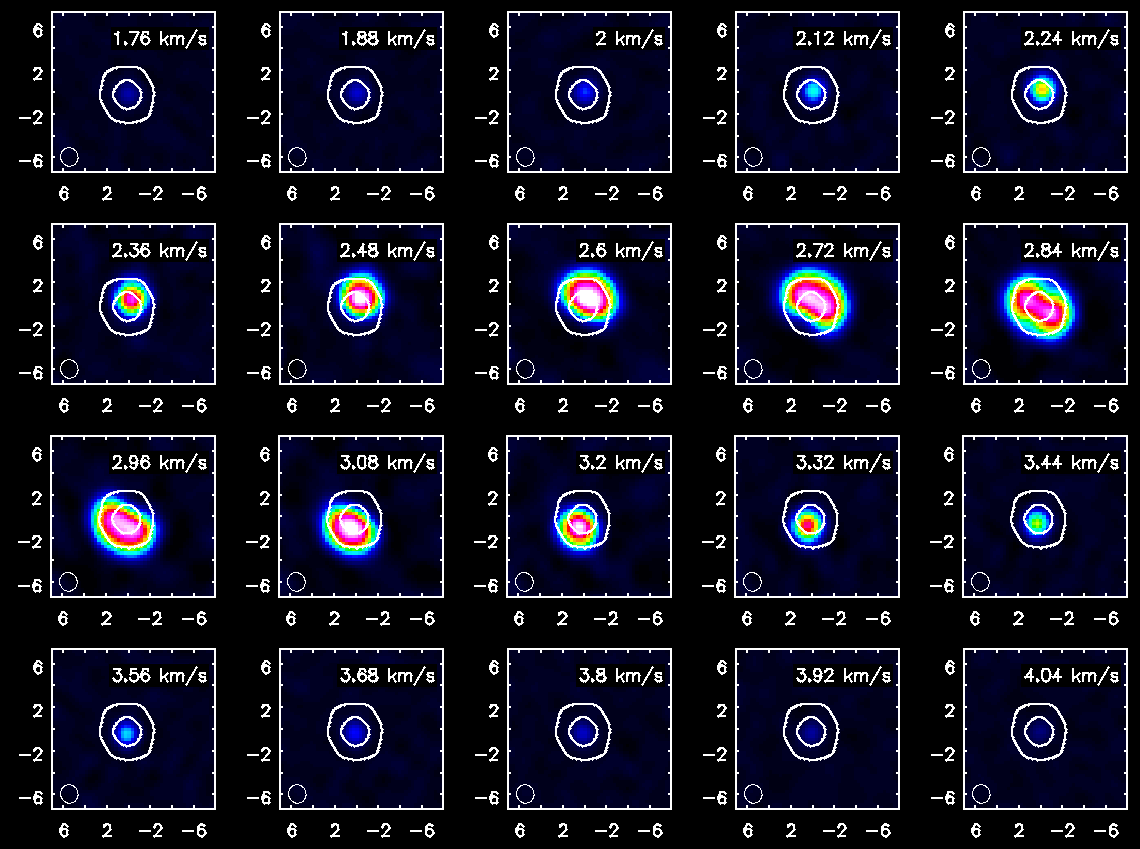
Channel maps of the HCO+(4-3) emission with white continuum contours at 3 and 100 sigma. The color intensity scale is the same as that used for CO(3-2).