First Look at Imaging: Difference between revisions
| Line 229: | Line 229: | ||
At this point, you can experiment with the {{plotms}} GUI. For example, it is often interesting and useful to examine the plot of amplitude vs. UV distance. Select the "Axes" tab, then set the X Axis to UVDist and the Data setting to "Amp". Under the "Page" tab, set Iteration Axis to "Field". Then click the Plot button. You can step through the different calibrators and science targets using the green arrow buttons at the bottom of the {{plotms}} GUI. | At this point, you can experiment with the {{plotms}} GUI. For example, it is often interesting and useful to examine the plot of amplitude vs. UV distance. Select the "Axes" tab, then set the X Axis to UVDist and the Data setting to "Amp". Under the "Page" tab, set Iteration Axis to "Field". Then click the Plot button. You can step through the different calibrators and science targets using the green arrow buttons at the bottom of the {{plotms}} GUI. | ||
== First Look at | == First Look at TCLEAN == | ||
Our secondary calibrator (also called the phase calibrator) is identified by field 3. Let's image this calibrator into an image file called "secondary." | Our secondary calibrator (also called the phase calibrator) is identified by field 3. Let's image this calibrator into an image file called "secondary." | ||
| Line 239: | Line 239: | ||
</source> | </source> | ||
Starting with CASA 4.7, the imaging capabilities of CLEAN have been refactored and improved in a task called TCLEAN | <pre style="background-color: #E0FFFF;"> | ||
Starting with CASA 4.7, the imaging capabilities of CLEAN have been refactored and improved in a task called TCLEAN, which is called by the user in largely the same way as clean. In this guide, we will use solely tclean. In the guide [https://casaguides.nrao.edu/index.php/TCLEAN_and_ALMA TCLEAN and ALMA], the differences in syntax between CLEAN and TCLEAN are made more explicit. For reference, legacy first look guides for older versions of CASA that use clean rather than tclean for imaging are kept here '''link TBD'''. | |||
</pre> | |||
Now we will use the task {{tclean}} to do the imaging. We set the image name to "secondary", specify that we want to image the data for field "3", and use multifrequency synthesis (specmode mfs) to make a single continuum image. Multifrequency synthesis combines data from all selected spectral channels into a single continuum image. Because the fractional bandwidth (delta nu/nu) is pretty small for this data set, we will not worry about the amplitude or structure of the source changing substantially with frequency. Therefore, we set nterms=1 and deconvolver=hogbom, telling TCLEAN that each deconvolved component has a single amplitude at all frequencies. We will image a single pointing, so we set gridder='standard'. We set the cell size to 0.1 arcseconds, which places ~5 pixels across a beam. You could figure out the beam size a priori, but it is often easier to just experiment with a quick imaging call and note the beam calculated by TCLEAN. In this case, TCLEAN reports a synthesized beam size of 0.58" x 0.51". As a rule of thumb, we'd like ~5 pixels across the smallest direction of our elliptical beam, so our choice of cell size is just right. We set the image size to 128x128 (but note that factors of 2 are not magic for CASA). This is enough to cover most of the primary beam, but we might want to image a wider field for a non-point source. The {{tclean}} task will start in interactive mode, which allows you to manually control the threshold, major cycles, and masking. | Now we will use the task {{tclean}} to do the imaging. We set the image name to "secondary", specify that we want to image the data for field "3", and use multifrequency synthesis (specmode mfs) to make a single continuum image. Multifrequency synthesis combines data from all selected spectral channels into a single continuum image. Because the fractional bandwidth (delta nu/nu) is pretty small for this data set, we will not worry about the amplitude or structure of the source changing substantially with frequency. Therefore, we set nterms=1 and deconvolver=hogbom, telling TCLEAN that each deconvolved component has a single amplitude at all frequencies. We will image a single pointing, so we set gridder='standard'. We set the cell size to 0.1 arcseconds, which places ~5 pixels across a beam. You could figure out the beam size a priori, but it is often easier to just experiment with a quick imaging call and note the beam calculated by TCLEAN. In this case, TCLEAN reports a synthesized beam size of 0.58" x 0.51". As a rule of thumb, we'd like ~5 pixels across the smallest direction of our elliptical beam, so our choice of cell size is just right. We set the image size to 128x128 (but note that factors of 2 are not magic for CASA). This is enough to cover most of the primary beam, but we might want to image a wider field for a non-point source. The {{tclean}} task will start in interactive mode, which allows you to manually control the threshold, major cycles, and masking. | ||
Revision as of 19:15, 30 October 2017
About this Guide
The purpose of this tutorial is to provide a first look at imaging ALMA data for those new to CASA.
Data delivered by ALMA is pre-calibrated either by ARC staff or by the ALMA calibration pipeline. The delivered data is ready for imaging. This tutorial demonstrates the basic procedures that will help you complete the imaging steps.
This guide covers the same material used in hands-on training sessions at NRAO Community Days events and ALMA Data Reduction tutorials presented by NAASC staff.
The "Imaging Tutorials for CASA Beginners" guides work for CASA versions 5.0, 5.1, and possibly earlier versions.
Getting CASA
If you do not already have CASA installed on your machine, you will have to download and install it.
Download and installation instructions are available here:
http://casa.nrao.edu/casa_obtaining.shtml
About the Sample Data: Continuum and N2H+ in TW Hydra
The data for this example comes from ALMA Project 2011.0.00340.S, "Searching for H2D+ in the disk of TW Hya v1.5", for which the PI is Chunhua Qi. Part of the data for this project has been published in Qi et al. 2013.
The original observation had three scientific objectives:
- Image the submm continuum structure in TW Hydra
- Image the H2D+ line structure (rest frequency 372.42138 GHz)
- Image the N2H+ line structure (rest frequency 372.67249 GHz)
The data used in our tutorial has already been calibrated. Furthermore, to make the data set more manageable, we have reduced it in size by averaging in time and frequency. Our goal will be to image the continuum emission and the N2H+ spectral line, which is bright and well suited for demonstrating the imaging techniques.
The spectral window we will image covers 234.375 MHz in bandwidth, and contains 384 channels spaced by 610 kHz. The data includes observations from 21 of the ALMA 12-m main array antennas, observed during Early Science Cycle 0.
Getting the Data
The data used in this tutorial is part of a larger data package used for NRAO calibration and imaging tutorials. If you plan on working through all of the "First Look" tutorials for CASA beginners, it is worth downloading the entire package now. If you plan on working through just this imaging tutorial then you can download a smaller data package (see below).
The complete package (4.1G) is available here:
https://bulk.cv.nrao.edu/synth/dred_workshops/sis14/ss_alma_data_v1p2.tar.gz
You can then unpack the data as follows:
# In bash
tar xvzf ss_alma_data_v1p2.tar.gz
For the imaging tutorial, we are interested in the data that have been pre-flagged and calibrated. The relevant data set is called "sis14_twhya_calibrated_flagged.ms". You should copy this data set into a new working directory, and you will start CASA from that directory.
# In bash
mkdir MyTutorial
cd MyTutorial
cp -r ../working_data/sis14_twhya_calibrated_flagged.ms .
Alternatively, if you do not want to download the full 4.1G data package, you can download just the smaller data file (600M) needed specifically for this tutorial as follows:
# In bash
mkdir MyTutorial
cd MyTutorial
wget -r -np -nH --cut-dirs=4 --reject "index.html*" https://bulk.cv.nrao.edu/synth/dred_workshops/sis14/working_data/sis14_twhya_calibrated_flagged.ms/
Starting CASA
At this point you should be working in a unix shell from the directory that contains the data. An "ls" command should show you the data file "sis14_twhya_calibrated_flagged.ms" in the current working directory.
# In bash
ls
And now you can start CASA:
# In bash
casa
You will notice that CASA opens a message window, for feedback from CASA tasks. The CASA prompt is presented in the terminal window. CASA is running on a python interpreter, so python syntax is valid here.
When you are finished working with your data, you can end your CASA session by typing "exit" at the command prompt.
Some CASA basics
A complete description of CASA is available in the cookbook. Here we will touch on a couple of useful concepts to get us started.
A few unix commands work directly in CASA, for example you can try the "ls" command and the "pwd" command:
# In CASA
ls
pwd
You can access all operating system commands as follows:
# In CASA
os.system('ls')
It is often convenient to store CASA instructions in a python script. If you have such a script, it can be executed using the following syntax:
# In CASA
# if you are running this as an extracted script, comment this line out
execfile('my_script.py')
Using tasks, and getting oriented with the data
The first step in all data reduction with CASA is to examine the header information and data structure for the data set using the listobs task. In CASA, there are two ways to execute tasks. You can either set the parameters one-by-one and then instruct CASA to "go" when you're ready, or you can execute the task with a single command. For example, here is how you can use the first method:
# In CASA
inp listobs
vis='sis14_twhya_calibrated_flagged.ms'
go
And here is an example of running the listobs task with a single call from the command line:
# In CASA
listobs(vis='sis14_twhya_calibrated_flagged.ms')
The output for listobs is printed to the CASA Message window.
Note that you can get help on any CASA task:
# In CASA
help listobs
The listobs output for our data set is as follows:
================================================================================
MeasurementSet Name: /users/rfriesen/casaguides/first_look_imaging/sis14_twhya_calibrated_flagged.ms MS Version 2
================================================================================
Observer: cqi Project: uid://A002/X327408/X6f
Observation: ALMA
Computing scan and subscan properties...
Data records: 80563 Total elapsed time = 5647.68 seconds
Observed from 19-Nov-2012/07:36:57.0 to 19-Nov-2012/09:11:04.7 (UTC)
ObservationID = 0 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent
19-Nov-2012/07:36:57.0 - 07:39:13.1 4 0 J0522-364 4200 [0] [6.05] [CALIBRATE_BANDPASS#ON_SOURCE,CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
07:44:45.2 - 07:47:01.2 7 2 Ceres 3800 [0] [6.05] [CALIBRATE_AMPLI#ON_SOURCE,CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
07:52:42.0 - 07:53:47.6 10 3 J1037-295 1900 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
07:56:23.5 - 08:02:11.3 12 5 TW Hya 8514 [0] [6.05] [OBSERVE_TARGET#ON_SOURCE]
08:04:36.3 - 08:05:41.9 14 3 J1037-295 1900 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
08:08:09.6 - 08:13:57.3 16 5 TW Hya 10360 [0] [6.05] [OBSERVE_TARGET#ON_SOURCE]
08:16:20.6 - 08:17:26.2 18 3 J1037-295 2100 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
08:19:53.9 - 08:25:41.7 20 5 TW Hya 10321 [0] [6.05] [OBSERVE_TARGET#ON_SOURCE]
08:28:17.1 - 08:29:22.6 22 3 J1037-295 2100 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
08:32:00.5 - 08:37:48.2 24 5 TW Hya 10324 [0] [6.05] [OBSERVE_TARGET#ON_SOURCE]
08:40:11.9 - 08:41:17.4 26 3 J1037-295 2100 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
08:43:45.6 - 08:49:33.4 28 5 TW Hya 9462 [0] [6.05] [OBSERVE_TARGET#ON_SOURCE]
08:51:57.1 - 08:53:02.6 30 3 J1037-295 1900 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
08:58:12.0 - 09:00:28.1 33 6 3c279 3402 [0] [6.05] [CALIBRATE_BANDPASS#ON_SOURCE,CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
09:01:35.7 - 09:02:41.2 34 3 J1037-295 1900 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
09:05:15.6 - 09:07:31.6 36 5 TW Hya 4180 [0] [6.05] [OBSERVE_TARGET#ON_SOURCE]
09:09:59.1 - 09:11:04.7 38 3 J1037-295 2100 [0] [6.05] [CALIBRATE_PHASE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE]
(nRows = Total number of rows per scan)
Fields: 5
ID Code Name RA Decl Epoch SrcId nRows
0 none J0522-364 05:22:57.984648 -36.27.30.85128 J2000 0 4200
2 none Ceres 06:10:15.950590 +23.22.06.90668 J2000 2 3800
3 none J1037-295 10:37:16.079736 -29.34.02.81316 J2000 3 16000
5 none TW Hya 11:01:51.796000 -34.42.17.36600 J2000 4 53161
6 none 3c279 12:56:11.166576 -05.47.21.52464 J2000 5 3402
Spectral Windows: (1 unique spectral windows and 1 unique polarization setups)
SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) BBC Num Corrs
0 ALMA_RB_07#BB_2#SW-01#FULL_RES 384 TOPO 372533.086 610.352 234375.0 372649.9688 2 XX YY
Sources: 5
ID Name SpwId RestFreq(MHz) SysVel(km/s)
0 J0522-364 0 - -
1 Ceres 0 - -
2 J1037-295 0 - -
3 TW Hya 0 - -
4 3c279 0 - -
Antennas: 21:
ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m)
East North Elevation x y z
1 DA42 A050 12.0 m -067.45.16.2 -22.53.29.3 43.0352 -744.9713 21.6702 2225079.880016 -5440041.377534 -2481724.598031
2 DA44 A068 12.0 m -067.45.20.6 -22.53.25.7 -82.4232 -631.7828 23.5810 2224981.097784 -5440131.250387 -2481621.066374
3 DA45 A070 12.0 m -067.45.11.9 -22.53.29.3 166.1833 -743.4934 19.8811 2225193.450167 -5439993.764157 -2481722.540534
4 DA46 A067 12.0 m -067.45.12.7 -22.53.27.2 142.4097 -678.7318 20.1280 2225181.070532 -5440026.290790 -2481662.975103
5 DA48 A046 12.0 m -067.45.17.0 -22.53.29.3 21.4267 -742.7987 21.6757 2225060.202580 -5440050.344436 -2481722.598651
6 DA49 A029 12.0 m -067.45.18.2 -22.53.25.8 -12.9134 -636.4552 22.1350 2225044.239583 -5440102.022535 -2481624.808405
7 DA50 A045 12.0 m -067.45.17.9 -22.53.30.1 -5.4183 -767.4398 22.6034 2225032.051652 -5440052.426015 -2481745.660003
9 DV02 A077 12.0 m -067.45.10.1 -22.53.25.9 217.6299 -637.5333 15.8376 2225255.259272 -5440008.987869 -2481623.352052
11 DV05 A082 12.0 m -067.45.08.3 -22.53.29.2 269.0433 -740.9521 15.7832 2225287.593766 -5439952.243679 -2481718.605314
12 DV06 A037 12.0 m -067.45.17.5 -22.53.28.8 6.7403 -727.3003 21.2086 2225048.729287 -5440061.085777 -2481708.139136
14 DV08 A021 12.0 m -067.45.17.2 -22.53.27.0 14.3196 -672.8108 21.3420 2225063.814715 -5440077.948261 -2481657.992572
15 DV10 A071 12.0 m -067.45.19.9 -22.53.23.5 -60.7887 -563.2541 23.3799 2225011.141945 -5440147.560932 -2481557.855663
16 DV13 A072 12.0 m -067.45.12.6 -22.53.24.0 147.1742 -580.5887 18.1825 2225199.254375 -5440058.161494 -2481571.803699
17 DV15 A074 12.0 m -067.45.12.1 -22.53.32.0 161.8159 -828.6196 18.7688 2225176.483514 -5439963.820451 -2481800.529842
18 DV16 A069 12.0 m -067.45.21.3 -22.53.30.2 -101.4797 -770.1047 23.2972 2224942.993176 -5440088.421459 -2481748.384855
19 DV17 A138 12.0 m -067.45.17.1 -22.53.34.4 19.1461 -901.2603 26.0137 2225036.269025 -5439997.853009 -2481870.267607
20 DV18 A053 12.0 m -067.45.17.3 -22.53.31.2 12.5939 -802.9941 21.5281 2225043.111690 -5440031.889497 -2481777.995870
21 DV19 A008 12.0 m -067.45.15.4 -22.53.26.8 67.5592 -667.6872 20.9574 2225113.709955 -5440059.310545 -2481653.122797
22 DV20 A020 12.0 m -067.45.17.8 -22.53.28.0 -2.9649 -703.4389 21.6629 2225043.419055 -5440073.737929 -2481686.333574
24 DV22 A011 12.0 m -067.45.14.4 -22.53.28.4 95.9131 -716.5005 21.0898 2225132.810230 -5440031.115405 -2481698.143589
25 DV23 A007 12.0 m -067.45.15.1 -22.53.27.3 74.0152 -681.2926 21.3231 2225117.809276 -5440052.280005 -2481665.799049
Inspecting the data
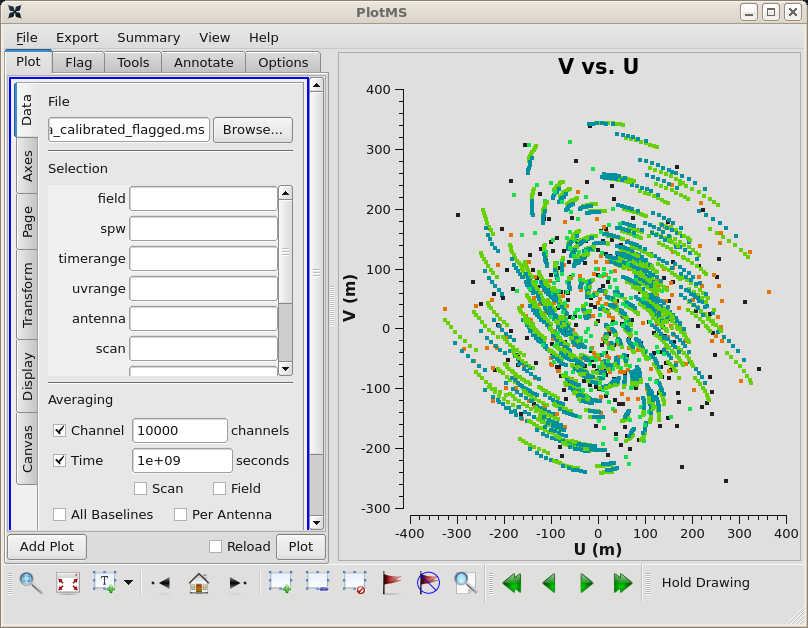
The plotms task is an important and versatile tool that allows the user to look at the UV data in a graphical manner. Let's start by plotting the UV coverage for our data.
# In CASA
plotms(vis='sis14_twhya_calibrated_flagged.ms', xaxis='u', yaxis='v', avgchannel='10000', avgspw=False, avgtime='1e9', avgscan=False, coloraxis="field", showgui=True)
<figure id="Imaging-tutorial-uv-coverage.png">
</figure>
You should see a GUI as in Figure 1. The colors in the UV coverage plot identify different observing targets.
At this point, you can experiment with the plotms GUI. For example, it is often interesting and useful to examine the plot of amplitude vs. UV distance. Select the "Axes" tab, then set the X Axis to UVDist and the Data setting to "Amp". Under the "Page" tab, set Iteration Axis to "Field". Then click the Plot button. You can step through the different calibrators and science targets using the green arrow buttons at the bottom of the plotms GUI.
First Look at TCLEAN
Our secondary calibrator (also called the phase calibrator) is identified by field 3. Let's image this calibrator into an image file called "secondary." First, in case you've run this task before, let's remove old versions of the image that use this name. (The ".*" is needed because imaging produces several files with the same root name).
# In CASA
os.system('rm -rf secondary.*')
Starting with CASA 4.7, the imaging capabilities of CLEAN have been refactored and improved in a task called TCLEAN, which is called by the user in largely the same way as clean. In this guide, we will use solely tclean. In the guide [https://casaguides.nrao.edu/index.php/TCLEAN_and_ALMA TCLEAN and ALMA], the differences in syntax between CLEAN and TCLEAN are made more explicit. For reference, legacy first look guides for older versions of CASA that use clean rather than tclean for imaging are kept here '''link TBD'''.
Now we will use the task tclean to do the imaging. We set the image name to "secondary", specify that we want to image the data for field "3", and use multifrequency synthesis (specmode mfs) to make a single continuum image. Multifrequency synthesis combines data from all selected spectral channels into a single continuum image. Because the fractional bandwidth (delta nu/nu) is pretty small for this data set, we will not worry about the amplitude or structure of the source changing substantially with frequency. Therefore, we set nterms=1 and deconvolver=hogbom, telling TCLEAN that each deconvolved component has a single amplitude at all frequencies. We will image a single pointing, so we set gridder='standard'. We set the cell size to 0.1 arcseconds, which places ~5 pixels across a beam. You could figure out the beam size a priori, but it is often easier to just experiment with a quick imaging call and note the beam calculated by TCLEAN. In this case, TCLEAN reports a synthesized beam size of 0.58" x 0.51". As a rule of thumb, we'd like ~5 pixels across the smallest direction of our elliptical beam, so our choice of cell size is just right. We set the image size to 128x128 (but note that factors of 2 are not magic for CASA). This is enough to cover most of the primary beam, but we might want to image a wider field for a non-point source. The tclean task will start in interactive mode, which allows you to manually control the threshold, major cycles, and masking.
# In CASA
tclean(vis='sis14_twhya_calibrated_flagged.ms',
imagename='secondary',
field='3',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[128,128],
cell=['0.1arcsec'],
weighting='natural',
threshold='0mJy',
interactive=True)
Note that with niter (the maximum number of iterations) = 0, TCLEAN is being told not to do any cleaning. If there are no clean components in the model -- for instance if this is the first invocation of TCLEAN on this field and spectral window for the given set of visibilities (MS) -- then what will be created is the dirty cube or image. If you run TCLEAN with niter=0 you will get the following warning message:
WARN task_tclean::SIImageStore::restore (file /var/rpmbuild/BUILD/casa-prerelease/casa-prerelease-4.7.0/code/synthesis/ImagerObjects/SIImageStore.cc, line 1764) Restoring with an empty model image. Only residuals will be processed to form the output restored image.
Run the command again, this time with niter set to some number:
# In CASA
tclean(vis='sis14_twhya_calibrated_flagged.ms',
imagename='secondary',
field='3',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[128,128],
cell=['0.1arcsec'],
weighting='natural',
threshold='0mJy',
niter=100,
interactive=True)
After running the clean task, you will be presented with the GUI as shown in Figure 2. In the CLEAN viewer, make sure that your buttons are set to add a new oval mask region. You may need to click on the icon showing the "R" in an oval. Remember that the secondary calibrator has been selected because it is a point source, so at this point you should see a point source in the middle of the field. Draw an oval mask around the emission region (just the central dot). Double click inside the oval and watch it turn white (See Figure 2). When setting the clean mask, you should aim to capture the real emission and not much else. In the "Next Action" section of the GUI, you will notice several control buttons. Hit the green circle button to begin the cleaning process. This will run a major cycle of cleaning and then return. After the first round of cleaning, the plot displays the residual emission after the major cycle. Look at those gorgeous residuals! When you are satisfied (or when CLEAN thinks the CLEANing has met the threshold, 0 mJy by default - meaning that it stops at the first negative), hit the red "X" and CLEAN will terminate. In this example, one round of cleaning is fine. For more complex targets you will need multiple rounds of cleaning, and it is possible to update and add new clean regions after each major cycle, based on the look of the residuals.
<figure id="Imaging-tutorial-secondary-clean.png">
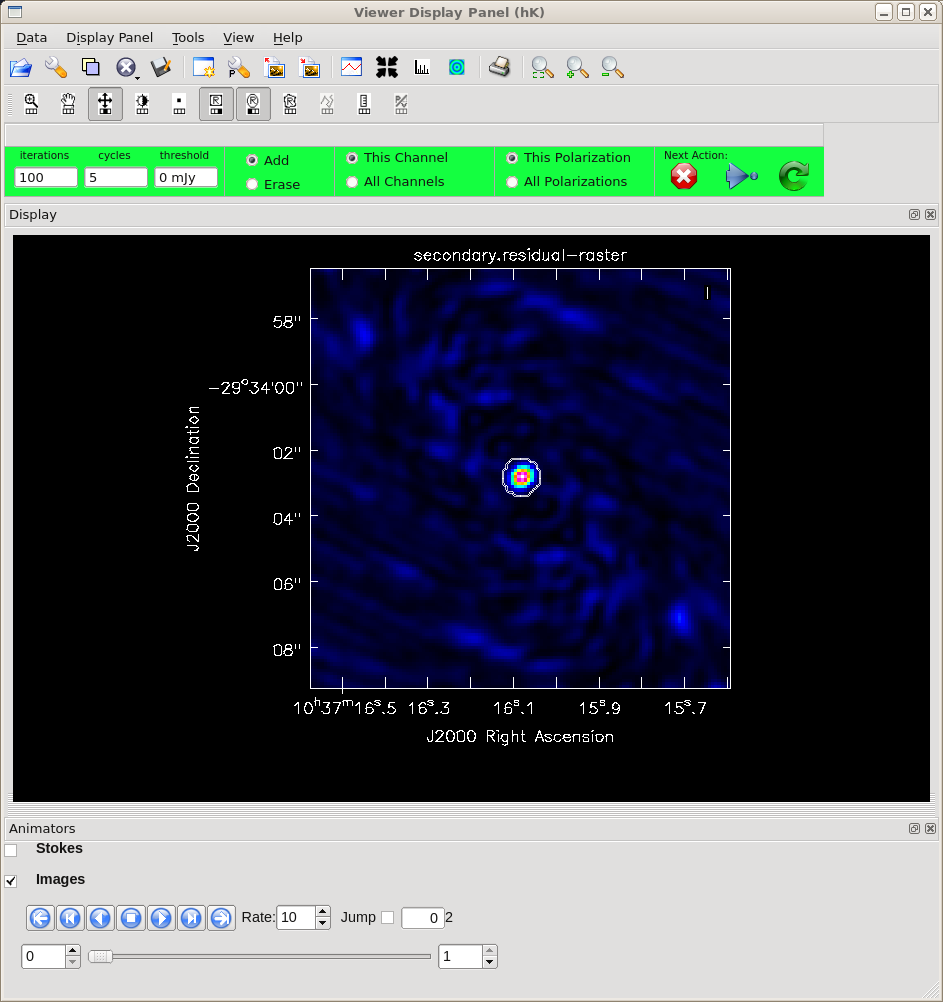
</figure>
Have a quick look at the files that CLEAN has created:
# In CASA
ls
The .image file is the final cleaned image. The .mask entry shows the clean mask, or the area that CLEANed, the .model is the set of modeled clean components used by CLEAN (in Jy/pixel), the .pb shows the primary beam response, the .residual shows what was left after you CLEANed (the "dirty" part of the final image), and the .psf file shows the synthesized beam. So much good stuff. You can look at all of these using the CASA viewer. From within CASA, the viewer can be started with "viewer()", or "imview()". You can also start the viewer as a stand-alone unix utility using the unix command "casaviewer". Here, let's examine the cleaned image:
# In CASA
imview("secondary.image")
Look at the other images now by loading them interactively using the viewer. Use the GUI to load several images, and then use the tape deck to control which image is actively displayed in the panel.
Experiment with TCLEAN
TCLEAN includes a lot of options. Remember, you can see the list of inputs for the task by typing "inp tclean". Now is a good time to get a feel for what these options can do. One option that is very commonly tweaked by the user is the weighting scheme used to grid the UV data into a fourier-plane image. This weighting was "natural" in the first example (by default). Try changing it to "briggs" here and try a few different values of the robust parameter. Pay attention to how the beam size changes, as well as the noise in the final image. You can check the noise level using the viewer -- draw a box on the image and double-right-click in the box to get image statistics in that region.
Remove old versions of the image in case you have run this before:
# In CASA
os.system('rm -rf secondary_robust.*')
Call TCLEAN with briggs weighting and robust = -1 :
# In CASA
tclean(vis='sis14_twhya_calibrated_flagged.ms',
imagename='secondary_robust',
field='3',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[128,128],
cell=['0.1arcsec'],
weighting='briggs',
robust=-1.0,
threshold='0mJy',
niter=100,
interactive=True)
Look at the results:
# In CASA
imview("secondary_robust.image")
Now is a good time to experiment a bit with TCLEAN - try imaging the other calibrators (fields 0 and 2) and making the image size and cell size larger and smaller.
For example, let's look at the marginally-resolved primary (flux) calibrator, Ceres (field 2):
# In CASA
os.system('rm -rf primary_robust.*')
tclean(vis='sis14_twhya_calibrated_flagged.ms',
imagename='primary_robust',
field='2',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[128,128],
cell=['0.1arcsec'],
weighting='natural',
threshold='0mJy',
niter=100,
interactive=True)
imview("primary_robust.image")
Notice that Ceres is somewhat resolved, leading to changing amplitude as a function of UV distance that is evident from the plotms plots.
If you try a really big pixel size you will see things break. It is recommended to have the pixel size small compared to the synthesized beam for TCLEANing purposes (TCLEAN quantizes the deconvolution in units of pixels). When the pixel size is big compared to the synthesized beam the imaging in general will degrade, even independent of TCLEANing.
# In CASA
os.system('rm -rf secondary_bigpix.*')
tclean(vis='sis14_twhya_calibrated_flagged.ms',
imagename='secondary_bigpix',
field='3',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[32,32],
cell=['0.5arcsec'],
weighting='natural',
threshold='0mJy',
niter=100,
interactive=True)
imview("secondary_bigpix.image")
To see the issues clearly here, compare the beam in this image to the one in the first image we made (with 5x smaller pixels).
ASIDE: See the effects of calibration and flagging
This section gives an aside intended to demonstrate the effect of calibration. If you are focused on learning only imaging, feel free to skip this section.
The data that we are imaging went through a careful and detailed calibration process. What effect did this actually have? Let's image the secondary calibrator with and without calibration and with and without flagging just to get an idea of how our processing changed the final image.
First you need to copy the uncalibrated data from the working directory. This step will differ depending on where you are storing the data. The net result we want is to have a copy of the uncalibrated data in the current working directory.
If you downloaded the full data package, copy the uncalibrated data to your current working directory.
# In CASA
os.system("rm -rf sis14_twhya_uncalibrated.ms")
os.system("cp -r ../working_data/sis14_twhya_uncalibrated.ms .")
If you downloaded only the calibrated data set, you can use "wget" again to download the uncalibrated data at this time:
# In bash
% wget -r -np -nH --cut-dirs=4 --reject "index.html*" https://bulk.cv.nrao.edu/synth/dred_workshops/sis14/working_data/sis14_twhya_uncalibrated.ms/
Now let's TCLEAN the uncalibrated data, again focusing on the secondary calibrator (field 3) and using the same calls as before.
# In CASA
os.system('rm -rf secondary_uncalibrated.*')
tclean(vis='sis14_twhya_uncalibrated.ms',
imagename='secondary_uncalibrated',
field='3',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[128,128],
cell=['0.1arcsec'],
weighting='natural',
threshold='0mJy',
niter=100,
interactive=True)
If you can find a source to TCLEAN then more power to you, but this is a mess. It's a good thing that we calibrated... In the raw (but still Tsys and WVR corrected) data you can see echos of the calibrator throughout the field, but the calibration is required to make the image coherent. Inspect the imaged uncalibrated data using the CASA viewer:
# In CASA
imview("secondary_uncalibrated.image")
Now let's see the effect that flagging had on the data. Copy the unflagged data from the working directory to our local directory:
# In CASA
os.system("rm -rf sis14_twhya_calibrated.ms")
os.system("cp -r ../working_data/sis14_twhya_calibrated.ms .")
or again, using wget:
# In bash
% wget -r -np -nH --cut-dirs=4 --reject "index.html*" https://bulk.cv.nrao.edu/synth/dred_workshops/sis14/working_data/sis14_twhya_calibrated.ms/
Now image the calibrated but unflagged data for the secondary calibrator using the same parameters as before.
# In CASA
os.system('rm -rf secondary_unflagged.*')
tclean(vis='sis14_twhya_calibrated.ms',
imagename='secondary_unflagged',
field='3',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[128,128],
cell=['0.1arcsec'],
weighting='natural',
threshold='0mJy',
niter=100,
interactive=True)
imview("secondary_unflagged.image")
In contrast to the uncalibrated data, the unflagged data are coherent, but they have clear artifacts in the residuals. Flagging has definitely improved the quality of the data, but in overall good quality data like we have here, we can see the target source.
Image the science target
Of course, the whole point of calibration is to calibrate the science data. Here, the TW Hydra observations use field 5, which we know from the output of listobs, above. As the final step in the basic imaging tutorial, let's now image the continuum in TW Hydra. First, we will "split out" the science data into its own data set. While not strictly necessary, this is a common step that makes managing the data easier. At the same time we will smooth (i.e. average) the data in frequency using width=10 to do the averaging. This reduces the data volume without losing much information, since we are really only interested in continuum imaging at this point. Although this data set was already designed to be manageable, this smoothing trick is good to keep in mind because ALMA can produce very large data sets.
# In CASA
os.system('rm -rf twhya_smoothed.ms')
split(vis='sis14_twhya_calibrated_flagged.ms', field='5', width='10', outputvis='twhya_smoothed.ms', datacolumn='data')
listobs('twhya_smoothed.ms')
Now make a continuum image of the split-out data. Notice that TW Hydra has now been re-labeled as field 0 in the new data set because we split out only that field. Again we will use the multifrequency synthesis mode ("mfs") and we will use both a somewhat smaller pixel size and a somewhat bigger image size than above (because TW Hydra is extended and the beam will be somewhat smaller due to our use of "briggs" weighting). Again, specify interactive mode and leave the threshold unset for the time being.
# In CASA
os.system('rm -rf twhya_cont.*')
tclean(vis='twhya_smoothed.ms',
imagename='twhya_cont',
field='0',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[250,250],
cell=['0.08arcsec'],
weighting='briggs',
robust=0.5,
threshold='0mJy',
niter=100,
interactive=True)
<figure id="Imaging-tutorial-twhydra-cont.png">
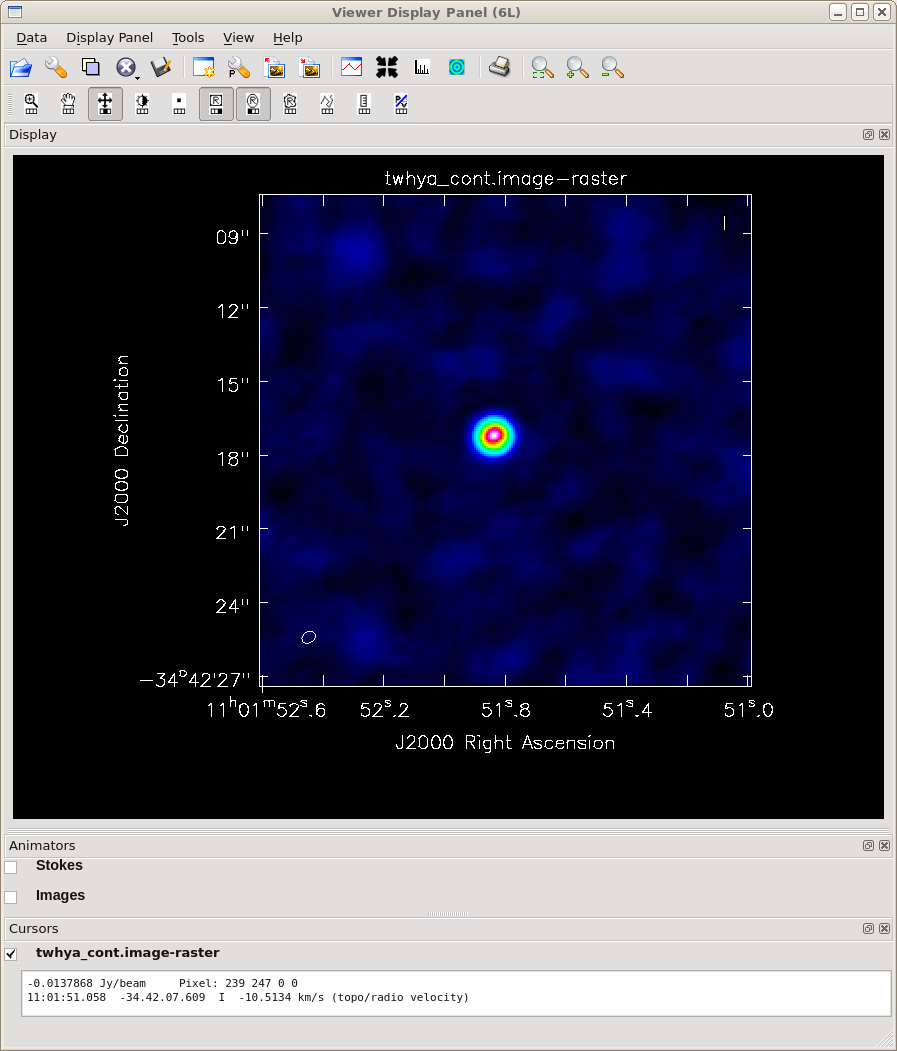
</figure>
Draw a clean mask around the visible emission using the mask tools and then TCLEAN until the emission from the TW Hydra disk is less than or comparable to the residuals around it. In this interactive mode of cleaning, you decide when TCLEAN should stop, and at that point you hit the red X. If instead you did want to clean automatically down to a threshold, you can specify a threshold that is a small multiple of the rms noise either in the call to TCLEAN above or by typing it in to the viewer window.
Have a look at the image - TW Hydra is very bright and you can see that it is extended relative to the size of the beam, which is represented by an oval at the bottom of the panel. The residuals aren't perfect, but we will improve them in subsequent lessons.
# In CASA
imview("twhya_cont.image")
Non-interactive clean
So far we have mostly followed an interactive process with TCLEAN. TCLEAN can also be set up to run without interactive guidance. The three main parameters to specify are the threshold at which to stop (when the maximum residual in the tclean region is lower than this threshold, TCLEAN stops), the mask (the region in which TCLEAN is willing to identify signal), and the maximum number of iterations. The max iterations is not strictly required and it is generally recommended that it be used as more of a failsafe. That is, set it to a number so high that if TCLEAN gets there something has gone wrong.
First let's take a stab at determining a clean mask. Look at the image you just made. All of the obvious emission is contained in a box that is bounded by pixel numbers in a range of something like (100,100) to (150,150). We'll set that box to be a mask using the "mask" parameter in the call to tclean. You could also set it by supplying a file (for example the one created from your earlier interactive version of tclean).
Now let's determine a stopping threshold for TCLEAN. Again, look at the previous image using imview and draw a box well away from the source to estimate the noise. Remember, you can right double-click in the box to get the RMS noise. We see something like ~7 mJy/beam. Set the threshold to be about twice this, ~15 mJy/beam. A clean threshold several times the rms noise is usually recommended to avoid adding false sources to the deconvolved image. That is, you do not want tclean to treat a random noise spike as a source and deconvolve it from the image. This can be particularly problematic if you are doing self-calibration (a later lesson). Finally set niter=5000, which is a lot of iterations. We expect TCLEAN to terminate before reaching this. For our purposes this is just a big number that's designed to keep TCLEAN from running forever.
# In CASA
os.system('rm -rf twhya_cont_auto.*')
tclean(vis='twhya_smoothed.ms',
imagename='twhya_cont_auto',
field='0',
spw='',
specmode='mfs',
gridder='standard',
nterms=1,
deconvolver='hogbom',
imsize=[250,250],
cell=['0.08arcsec'],
mask='box [ [ 100pix , 100pix] , [150pix, 150pix ] ]',
weighting='briggs',
robust=0.5,
threshold='15mJy',
niter=5000,
interactive=False)
imview('twhya_cont_auto.image')
Look ma, no hands!
This noninteractive mode can save you a lot of time and has the advantage of being very reproducible. Note that you also have a "hybrid" mode available by starting the TCLEAN process with interactive=True but then hitting the blue arrow button in the top right corner. This tells TCLEAN to proceed until it hits the maximum number of iterations or the threshold. This combination mode is nice because you can manually draw the mask used to tclean. Note that you can also manually set both the threshold and the maximum number of iterations (which is the product of the number of major cycles and the iterations per cycle) in the viewer GUI. Note, however, that best practice for an image with uncertain calibration and especially one with a bright source, is to clean interactively at least the first time. In the case where an image may be "dynamic range limited" (i.e., the quality is set by the accuracy of calibration and deconvolution) it can be hard to predict the correct threshold.
As the cleaning cycles proceed, the residuals in the image should decrease; notice that tclean will inform the user if the peak residual within the masked region increases (by any amount) with "WARN" messages like these: does this still happen? no warnings in 5.1
WARN task_tclean::SIIterBot_state::cleanComplete (file /var/rpmbuild/BUILD/casa-prerelease/casa-prerelease-4.7.0/code/synthesis/ImagerObjects/SIIterBot.cc, line 142) Peak residual increased from 0.024549 to 0.0245551 WARN task_tclean::SIIterBot_state::cleanComplete (file /var/rpmbuild/BUILD/casa-prerelease/casa-prerelease-4.7.0/code/synthesis/ImagerObjects/SIIterBot.cc, line 142) Peak residual increased from 0.0148844 to 0.0148845
Primary beam correction
An important subtlety of TCLEAN (and CLEAN) is that by default the image produced by TCLEAN is not corrected for the primary beam (the field of view) of the individual dishes in the array. The primary beam response is typically a Gaussian with value 1 at the center of the field. To form an astronomically correct image of the sky, the output of TCLEAN needs to be divided by this primary beam (or, in the case of mosaics, the combination of primary beam patterns used to make the mosaic). Fortunately, CASA stores the primary beam information needed to make this correction in an image file with a ".pb" extension.
The CASA task impbcor can be used to combine the .pb image with the output image from TCLEAN to produce a primary-beam corrected image.
First remove the old primary beam corrected image if it exists
# In CASA
os.system('rm -rf twhya_cont.pbcor.image')
Now correct the image
# In CASA
impbcor(imagename='twhya_cont.image',
pbimage='twhya_cont.pb',
outfile='twhya_cont.pbcor.image')
Inspect the output image
# In CASA
imview('twhya_cont.pbcor.image')
It's often very convenient to work in images before primary beam correction because the noise is the same across the field (e.g., this is a clean data set to search for signal) but it's very important to remember to apply this correction before calculating fluxes or intensities for science.