Guide To Processing ALMA Data for Cycle 0: Difference between revisions
No edit summary |
|||
| Line 159: | Line 159: | ||
In most cases, users should not need to redo the calibration. However, you should consult [https://help.almascience.org/index.php?/na/Knowledgebase/Article/View/162/ this knowledgebase article] for suggestions on when recalibration may be beneficial. If you do not need to redo calibration, proceed to the section on Data Concatenation. | In most cases, users should not need to redo the calibration. However, you should consult [https://help.almascience.org/index.php?/na/Knowledgebase/Article/View/162/ this knowledgebase article] for suggestions on when recalibration may be beneficial. If you do not need to redo calibration, proceed to the section on Data Concatenation. | ||
The data package includes a calibration script for each execution of the scheduling block. This script performs all the initial calibrations required prior to imaging the data set. The calibration script is the exact version used to generate the packaged, calibrated data | The data package includes a calibration script for each execution of the scheduling block. This script performs all the initial calibrations required prior to imaging the data set. The calibration script is the exact version used to generate the packaged, calibrated data, and the user can use that script and tweak it However, for Cycle 0 data sets the script cannot be applied directly by the user to re-calibrate. There are a couple of considerations which need to be taken into account before using the packaged data reduction script. | ||
# The "raw" data supplied with the data package have had several a priori calibrations applied. These include the calibration steps labeled 0-6 in the packaged scripts. So the user should begin with the step labeled "7" in the packaged data reduction script. | # The "raw" data supplied with the data package have had several a priori calibrations applied. These include the calibration steps labeled 0-6 in the packaged scripts. So the user should begin with the step labeled "7" in the packaged data reduction script (or "8", if using a CASA 4.2 script). This is done, in the CASA, by precising | ||
<source lang="python"> | |||
# In CASA | |||
mysteps=[7,8,9,10,11,12,13,14,15] | |||
execfile('scriptForCalibration_3.4.py') | |||
</source> | |||
# The directory structure provided in the Cycle 0 data distribution does not quite match the structure expected by the calibration scripts. | # The directory structure provided in the Cycle 0 data distribution does not quite match the structure expected by the calibration scripts. | ||
# The data processing script was generated using CASA 3.4, but the users are recommended to use CASA 4.2 for all processing. There are several differences between these versions. The CASA 3.4 script should be used, therefore, only as a guide to what needs to be done, and in what order. But the script must be modified to work under CASA 4.2. | # The data processing script was generated using CASA 3.4, but the users are recommended to use CASA 4.2 for all processing. There are several differences between these versions. The CASA 3.4 script should be used, therefore, only as a guide to what needs to be done, and in what order. But the script must be modified to work under CASA 4.2. | ||
| Line 168: | Line 173: | ||
In the '''raw''' data directory are 3 measurement sets that are ready for calibration. [Here] are 3 calibration scripts updated to CASA 4.2, that can be used to calibrate these data sets. They can be run in CASA 4.2 as follows. (Start from the '''raw''' directory.) | In the '''raw''' data directory are 3 measurement sets that are ready for calibration. [Here] are 3 calibration scripts updated to CASA 4.2, that can be used to calibrate these data sets. They can be run in CASA 4.2 as follows. (Start from the '''raw''' directory.) | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
thesteps = [9,10,11,12,13,14,15,16,17,18 | thesteps = [9,10,11,12,13,14,15,16,17,18] | ||
execfile('uid___A002_X554543_X207.ms.scriptForCalibration_4.2.py') | execfile('uid___A002_X554543_X207.ms.scriptForCalibration_4.2.py') | ||
execfile('uid___A002_X554543_X3d0.ms.scriptForCalibration_4.2.py') | execfile('uid___A002_X554543_X3d0.ms.scriptForCalibration_4.2.py') | ||
Revision as of 16:52, 26 March 2014
Using the ALMA Data Archive for Cycle 0 Data
(rename the page with this title, when finished)
About this Guide, and Cycle 0 ALMA Data
This guide describes steps that you can use to make science-ready images from Cycle 0 ALMA data available from the ALMA data archive. We begin with the process of locating and downloading the data from the public archive.
This guide is explicitly written for Cycle 0 data. Data for Cycle 1 and beyond will be delivered in a different format and will be described in a separate guide.
For Cycle 0, ALMA data is delivered with a set of calibrated data files and a sample of reference images. The data were calibrated and imaged by an ALMA scientist at one of the ALMA Regional Centers (ARCs). The scientist developed CASA scripts to complete the calibration and imaging. Cycle 0 data packages also include fully calibrated data sets, and the user can start with these data and proceed directly to imaging steps. In many cases, the imaging can be dramatically improved with "self-calibration" on the science target. Self-calibration is the process of using the detected signal in the target source, itself, to tune the phase (and perhaps amplitude) calibrations, as a function of time.
Typically, users interested in making science-ready images with Cycle 0 data from the ALMA archive will take the following steps:
- Download the data from the Archive
- Inspect the Quality Assurance plots and data files
- Inspect the reference images supplied with the data package
- Combine the calibrated data sets into a single calibrated measurement set
- Self-calibrate and image the combined data set
- Generate moment maps and other analysis products
In most cases, users will not need to modify the calibration supplied in the data package. But in some cases, users may wish to review the calibration steps in detail, make modifications to the calibration scripts, and generate new calibrated data sets.
About the Sample Data: H2D+ and N2H+ in TW Hya
The data for this example comes from ALMA Project 2011.0.00340.S, "Searching for H2D+ in the disk of TW Hya v1.5", for which the PI is Chunhua Qi. Part of the data for this project has been published in Qi et al. 2013.
This observation has three scientific objectives:
- Image the submm continuum structure in TW Hya
- Image the H2D+ line structure (rest frequency 372.42138 GHz)
- Image the N2H+ line structure (rest frequency 372.67249 GHz)
Eventually we will see that the data is distilled to 4 "spectral windows" used for science. The N2H+ line is in one spectral window, the H2D+ is in another window, and the other two windows contribute to the continuum observation. The best continuum map will be generated from the line-free channels of all 4 spectral windows.
Each spectral window covers 234.375 MHz in bandwidth, and the raw data contain 3840 channels, spaced by 61 kHz. Information on the spectral configuration and targets observed must be gleaned from the data themselves. In CASA, you can explore the contents of a data set using the listobs command.
What other data sets are available?
A Delivery List of publicly available data sets is provided on the ALMA Science Portal, in the "Data" tab.
Prerequisites : Computing Requirements
ALMA data sets can be very large and require significant computing resources for efficient processing. The data set used in this example begins with a download of 176 GB of data. For a recommendation on computing resources needed to process ALMA data, check here. Those who do not have sufficient computing power may wish to arrange a visit to one of the ARCs to use the computing facilities at these sites. To arrange a visit to an ARC, submit a ticket to the ALMA Helpdesk. Remote use of ARC computing facilities is not available at this time.
Getting the Data: The ALMA Data Archive
The ALMA data archive is part of the ALMA Science Portal. A copy of the archive is stored at each of the ARCs, and you can connect to the nearest archive through these links:
<figure id="Archive_interface.png">
</figure>
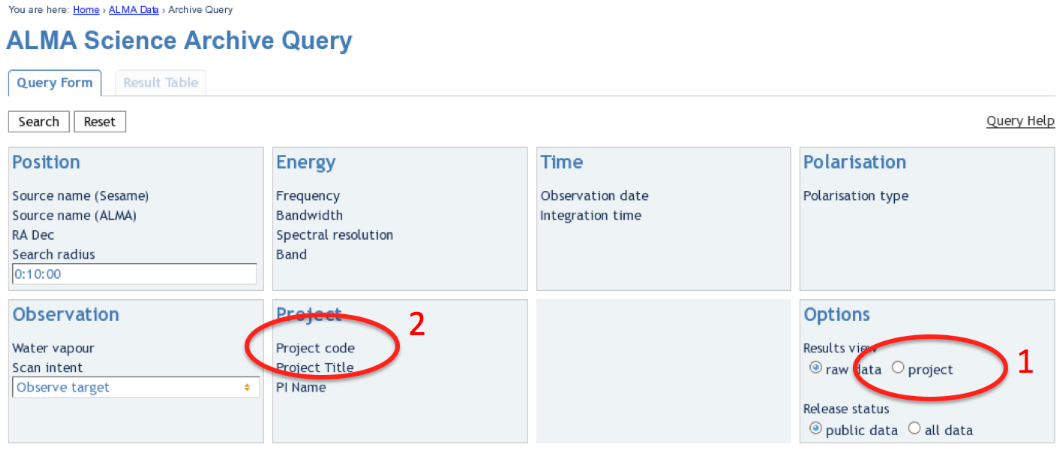
Upon entry into the ALMA Archive Query page, set the "Results View" option to "project" (see the red highlight #1 in Figure 1) and specify the Project Code to 2011.0.00340.S (red highlight #2). Note, if you leave the "Results View" set to "raw data", you will be presented with three rows of data sets in the results page. These correspond to three executions of the observing script. In fact, for Cycle 0 data these rows contains copies of the same data set, so use care not to download the (large!) data set three times. By setting "Results View" to "project", you see just one entry, and that is the one you'd like to download.
You can download the data through the Archive GUI. For more control over the download process, you can use the Unix shell script provided on the Request Handler page. This script has a name like "downloadRequest84998259script.sh". You need to put this file into a directory that contains ample disk space, and execute the script in your shell. For example, in bash:
# In bash
% chmod +x downloadRequest84998259script.sh
% ./downloadRequest84998259script.sh
Unpacking the data
The data you have downloaded for this particular project includes 17 tar files. Unpack these using the following command:
# In bash
% for i in $(ls *.tar); do echo 'Untarring ' $i; tar xvf $i; done
At this point you will have a directory called "2011.0.00340.S" with the full data distribution.
Overview of Delivered Data and Products
To get to the top-level directory that contains the data package, do:
# In bash
% cd 2011.0.00340.S/sg_ouss_id/group_ouss_id/member_ouss_2012-12-05_id
Here you will find the following entries:
# In bash
% ls
calibrated calibration log product qa raw README script
These directories are standard for all cycle 0 data deliveries. The README file describes the files in the distribution and includes notes from the ALMA scientist who performed the initial calibration and imaging.
In our example, the directories contain the following files:
-- calibrated/ |-- uid___A002_X554543_X207.ms.split.cal/ |-- uid___A002_X554543_X3d0.ms.split.cal/ |-- uid___A002_X554543_X667.ms.split.cal/ -- calibration/ |-- uid___A002_X554543_X207.calibration/ |-- uid___A002_X554543_X207.calibration.plots/ |-- uid___A002_X554543_X3d0.calibration/ |-- uid___A002_X554543_X3d0.calibration.plots/ |-- uid___A002_X554543_X667.calibration/ |-- uid___A002_X554543_X667.calibration.plots/ -- log/ |-- uid___A002_X554543_X207.calibration.log |-- uid___A002_X554543_X3d0.calibration.log |-- uid___A002_X554543_X667.calibration.log |-- Imaging.log |-- 340.log -- product/ |-- TWHya.continuum.fits |-- TWHya.N2H+.fits |-- TWHya.continuum.mask/ |-- TWHya.H2D+.mask/ |-- TWHya.N2H+.mask/ -- qa/ |-- uid___A002_X554543_X207__textfile.txt |-- uid___A002_X554543_X207__qa2_part1.png |-- uid___A002_X554543_X207__qa2_part2.png |-- uid___A002_X554543_X207__qa2_part3.png |-- uid___A002_X554543_X3d0__textfile.txt |-- uid___A002_X554543_X3d0__qa2_part1.png |-- uid___A002_X554543_X3d0__qa2_part2.png |-- uid___A002_X554543_X3d0__qa2_part3.png |-- uid___A002_X554543_X667__textfile.txt |-- uid___A002_X554543_X667__qa2_part1.png |-- uid___A002_X554543_X667__qa2_part2.png |-- uid___A002_X554543_X667__qa2_part3.png -- raw/ |-- uid___A002_X554543_X207.ms.split/ |-- uid___A002_X554543_X3d0.ms.split/ |-- uid___A002_X554543_X667.ms.split/ -- script/ |-- uid___A002_X554543_X207.ms.scriptForCalibration.py |-- uid___A002_X554543_X3d0.ms.scriptForCalibration.py |-- uid___A002_X554543_X667.ms.scriptForCalibration.py |-- scriptForImaging.py |-- import_data.py |-- scriptForFluxCalibration.py
- calibrated: This directory contains a calibrated measurement set for each of the execution blocks in the project. In our case, there are three. These could be imaged separately, or combined to make a single calibrated measurement set. In this guide we will combine the three executions prior to imaging.
- calibration: Auxiliary data files generated in the calibration process.
- The "calibration.plots" directories contain (a few hundred) plots generated during the calibration process. These can be useful for the expert user to assess the quality of the calibration at each step.
- log: Log files from the CASA sessions used by the ALMA scientist to calibrate and image the data. These are provided for reference.
- product: The final data products from the calibration and imaging process. The directory contains "reference" images that are used to determine the quality of the observation, but they are not necessarily science-ready. The data files here can used for initial inspections.
- qa: The result of standardized Quality Assessment tests. The data from each scheduling block goes through such a quality assessment, and all data delivered to the public ALMA Archive have passed the quality assessment. It is worthwhile to review the plots and text files contained here. You will find plots of the antenna configuration, UV coverage, calibration results, Tsys, and so on.
- raw: The "raw" data files. If you would like to tune or refine the calibration, these files would be your starting point. In fact, these data files do have certain a priori calibrations already applied, amounting to steps 0-6 of the calibration scripts provided in the data package. These steps include a priori flagging and application of WVR, Tsys, and antenna position corrections.
- script: These are the scripts developed and applied by the ALMA scientist, to calibrate the data and generate reference images. These scripts cannot be applied directly to the raw data provided in this data distribution, but they do serve as a valuable reference to see the steps that need to be taken to re-calibrate the data, if you so choose.
What if I need to redo the Calibration?
In most cases, users should not need to redo the calibration. However, you should consult this knowledgebase article for suggestions on when recalibration may be beneficial. If you do not need to redo calibration, proceed to the section on Data Concatenation.
The data package includes a calibration script for each execution of the scheduling block. This script performs all the initial calibrations required prior to imaging the data set. The calibration script is the exact version used to generate the packaged, calibrated data, and the user can use that script and tweak it However, for Cycle 0 data sets the script cannot be applied directly by the user to re-calibrate. There are a couple of considerations which need to be taken into account before using the packaged data reduction script.
- The "raw" data supplied with the data package have had several a priori calibrations applied. These include the calibration steps labeled 0-6 in the packaged scripts. So the user should begin with the step labeled "7" in the packaged data reduction script (or "8", if using a CASA 4.2 script). This is done, in the CASA, by precising
# In CASA
mysteps=[7,8,9,10,11,12,13,14,15]
execfile('scriptForCalibration_3.4.py')
- The directory structure provided in the Cycle 0 data distribution does not quite match the structure expected by the calibration scripts.
- The data processing script was generated using CASA 3.4, but the users are recommended to use CASA 4.2 for all processing. There are several differences between these versions. The CASA 3.4 script should be used, therefore, only as a guide to what needs to be done, and in what order. But the script must be modified to work under CASA 4.2.
Updating a script to work with CASA 4.2
In the raw data directory are 3 measurement sets that are ready for calibration. [Here] are 3 calibration scripts updated to CASA 4.2, that can be used to calibrate these data sets. They can be run in CASA 4.2 as follows. (Start from the raw directory.)
# In CASA
thesteps = [9,10,11,12,13,14,15,16,17,18]
execfile('uid___A002_X554543_X207.ms.scriptForCalibration_4.2.py')
execfile('uid___A002_X554543_X3d0.ms.scriptForCalibration_4.2.py')
execfile('uid___A002_X554543_X667.ms.scriptForCalibration_4.2.py')
execfile('scriptForFluxCalibration_4.2.py')
Data Concatenation
The data package includes a fully calibrated data set for each of the execution blocks. Each data set has been calibrated first according to the steps in the scriptForCalibration scripts, and subsequently by the steps listed in the scriptForFluxCalibration.py script. The data delivery package does not include a concatenated measurement set, so we will do the concatenation here, then proceed to imaging. Let's make a new directory for imaging, and do all subsequent work from there.
# In bash
mkdir Imaging
cd Imaging
casapy
and then, in CASA:
# In CASA
concat(vis = ['../calibrated/uid___A002_X554543_X207.ms.split.cal', '../calibrated/uid___A002_X554543_X3d0.ms.split.cal',
'../calibrated/uid___A002_X554543_X667.ms.split.cal'], concatvis = 'calibrated.ms')
This operation could take an hour or longer. When completed, we have created a calibrated measurement set calibrated.ms, which is ready for imaging.
Imaging
A first step is to list the contents of the calibarated.ms measurement set.
# In CASA
listobs(vis='calibrated.ms',listfile='calibrated.listobs')
The text file calibrated.listobs now contains a summary of the contents of this data file. The listobs output includes the following section:
Spectral Windows: (4 unique spectral windows and 1 unique polarization setups) SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) BBC Num Corrs 0 ALMA_RB_07#BB_1#SW-01#FULL_RES 3840 TOPO 372286.368 61.035 234375.0 1 XX YY 1 ALMA_RB_07#BB_2#SW-01#FULL_RES 3840 TOPO 372532.812 61.035 234375.0 2 XX YY 2 ALMA_RB_07#BB_3#SW-01#FULL_RES 3840 TOPO 358727.946 -61.035 234375.0 3 XX YY 3 ALMA_RB_07#BB_4#SW-01#FULL_RES 3840 TOPO 358040.598 -61.035 234375.0 4 XX YY
We see that the data file contains 4 spectral windows. The H2D+ data are contained in SpwID=0 (rest frequency of H2D+: 372.42138 GHz) and the N2H+ data are contained in SpwID=1 (rest frequency of N2H+: 372.67249 GHz). Note that the Ch0 column gives the sky frequency at channel 0. To get central frequencies for the spectral windows, add 117.1875 MHz for Spectral Window IDs 0 and 1. Subtract 117.1875 MHz for IDs 2 and 3. (Note the channel width for these IDs is negative, indicating the frequencies increase in the opposite direction as SpwID 0 and 1.
Our strategy will be to image and self-calibrate the continuum data first. We will then apply the self-cal solutions to each of the two line data files, and image those second. In general terms, we will follow the steps outlines in the Science Verification CASAguide on TW Hya. You can also refer to the ScriptForImaging.py script included with the data package.
Other data sets may require different strategies. This strategy works best in this case because the continuum is bright, and suitable for self-calibration. A data set with a bright line may benefit from self-calibration on the line, instead of the continuum.
Continuum Imaging
First we will split off the TW Hya data and average the data over 100 channels, eliminating a few channels at each edge prior to averaging. This will generate a data set suitable for continuum imaging. The final data set has 6.1 MHz channels, sufficient to allow efficient cleaning with the "Multi-Frequency Synthesis" algorithm used by the CASA task clean.
# In CASA
split(vis='calibrated.ms', outputvis='TWHya_cont.ms', datacolumn='data', keepflags=T, field='TW Hya', spw='0~3:21~3820', width=100)
listobs(vis='TWHya_cont.ms', listfile='TWHya_cont.listobs')
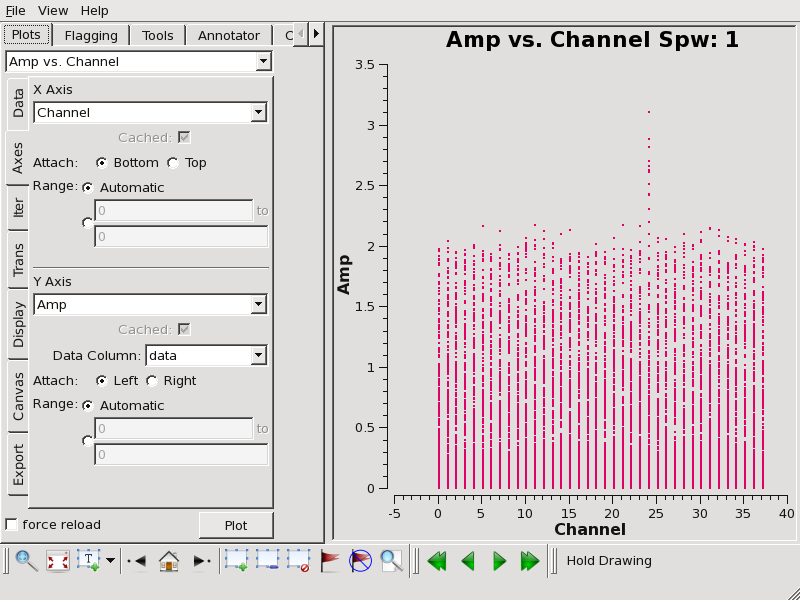
Now plot amplitude vs. channel with plotms to see what needs to be flagged.
# In CASA
plotms(vis='TWHya_cont.ms',spw='0~3',xaxis='channel',yaxis='amp',
avgtime='1e8',avgscan=T,coloraxis='spw',iteraxis='spw',xselfscale=T)
<figure id="Plotms_amp.png">
</figure>
We flag the line emission as follows:
# In CASA
flagdata(vis='TWHya_cont.ms', mode='manual', spw='1:24~24')
Next we will generate a first-pass continuum image. Note the mode "mfs" will generate a single continuum image from all the data.
# In CASA
os.system('rm -rf TWHya.continuum*')
clean(vis = 'TWHya_cont.ms',
imagename = 'TWHya.continuum',
field = '1', # TW Hya
spw = '0~3',
mode = 'mfs',
imagermode = 'csclean',
interactive = T,
imsize = [300, 300],
cell = '0.1arcsec',
phasecenter = 1,
weighting = 'briggs',
robust = 0.5)
Note that at ~370 GHz the ALMA primary beam has a width of ~ 20 arcseconds FWHM. We want to image an area at least as large as that. Also, for this data set the synthesized beam is about 0.49 x 0.46 arcseconds. We have selected a cell size of 0.1 arcsec to fully sample the synthesized beam.
We'd like to do some cleaning to achieve our best image. In clean, the multifrequency synthesis mode grids each spectral channel independently. Since the uv-spacing is a function of frequency, this will actually achieve greater uv-coverage than if all the channels and spectral windows had been averaged in advance.
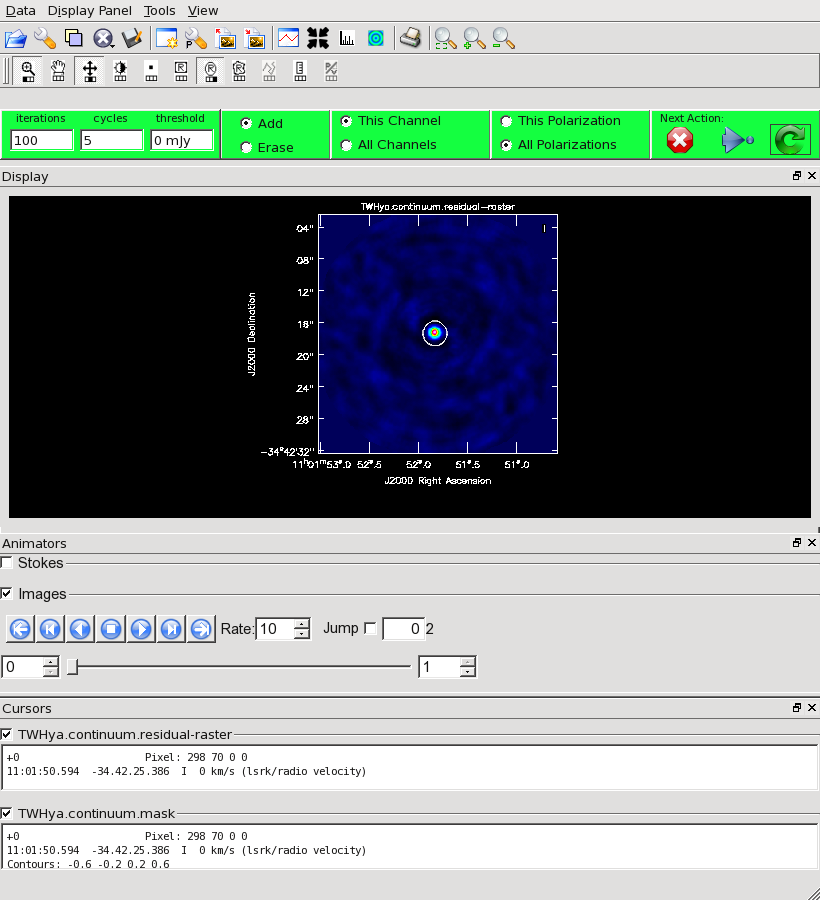
Our initial cleaned model will be used as the image model in the next self-calibration step. It is essential that only "real" signal be included in the image model, so we will clean the image conservatively. We set interactive=T to make the clean mask interactively. This also allows us to stop clean when the signal from the source starts to approach the surrounding "noise" areas in the residual map.
<figure id="Cont_clean1.png">
</figure>
Once the Clean Viewer opens, click on the ellipse tool with your left mouse button, and draw a circle around the continuum emission with the left mouse button. Double click inside the region with the left mouse button when you are happy with it. It will turn from green to white when the clean mask is accepted. To begin the clean use the green arrow in the "Next action" section on the Viewer Display GUI.
The red X will stop clean, the blue arrow will clean non-interactively until reaching the number of iterations requested (niter) or the flux density threshold (whichever comes first), and the green circle arrow will clean until it reaches the "iterations" parameter on the left side of the green area.
You should chose the green circle arrow. It will clean 100 iterations (as shown in the GUI) and then show the residual image. If necessary you can adjust the color scale to examine the residuals more clearly. The middle mouse button is initially assigned to the "plus" icon, which adjusts the colorscale. Also, note in the logger that clean reports the amount of clean flux recovered so far, the min/max residuals, and the synthesized beam.
In this example we will clean 500 iterations by clicking the green arrow five times, without changing the clean mask. After this we have cleaned about 1.5 Jy and we have residuals of about 5-7 mJy. Click the red and white "X" icon to conclude the cleaning process.
There are a few points to note:
- Real emission will not get "cleaned" if you do not put a clean box on it.
- Conversely, if you put a clean box on a region that contains no real emission and clean deeply, you can amplify that feature.
- Thus, it's best to be conservative. Stop cleaning when the residuals inside the clean box approach those outside it. Start with a small clean box surrounding what is clearly real emission, and add to it after some iterations, if necessary. You can generally see fainter "real" features in the residuals.
- If you start an interactive clean and then do not make a mask, the algorithm will clean nothing. There is no default mask.
clean will generate several files:
- TWHya.continuum.flux # the effective primary beam response (e.g. for pbcor)
- TWHya.continuum.flux.pbcoverage # the primary beam coverage (ftmachine=’mosaic’ only)
- TWHya.continuum.image # the final restored image
- TWHya.continuum.mask # the clean mask
- TWHya.continuum.model # the model image
- TWHya.continuum.psf # the synthesized (dirty) beam
- TWHya.continuum.residual # the residual image
To examine the cleaned image with the CASA viewer, use:
# In CASA
viewer('TWHya.continuum.image')
First cycle of self-calibration: phase only
Next we can use the clean model to self-calibrate the uv-data. The process of self-calibration tries to adjust the data to better match the model you give it. That is why it is important to make the model as good as you can. clean places a uv-model of the resulting image in the file TWHydra_cont.model. This model can then be used to self-calibrate the data using gaincal. We use gaintype='T' to tell it to average the polarizations before deriving phase solutions (to improve sensitivity by a factor of sqrt(2)). The "solution interval" (solint) is critical and may require some experimentation to get optimal results. From listobs we see that the integration time of these data is 6.05 seconds, so we want to select a solution interval that is at least this long, and is matched to the expected coherence time of the atmosphere, while maintaining adequate S/N. We can use solint=30s as a starting point. It is often best to start with a larger solint and then work your way down to the integration time if the S/N of the data permits. For the reasons described in the TWHydraBand7_Calibration_4.2 tutorial, we want to determine the phases before tackling amplitudes. Se we set calmode='p'.
# In CASA
gaincal(vis='TWHya_cont.ms',caltable='self_1.pcal',
solint='30s',combine='',gaintype='T',
refant='DV22',spw='',minblperant=4,
calmode='p',minsnr=2)
<figure id="Self_1_phase.png">
</figure>
We are using combine=' ' to get independent solutions for each spw. If S/N is insufficient to get independent solutions, you can try combining spectral windows with combine='spw'. We have plenty of S/N in these data so we keep them separate.
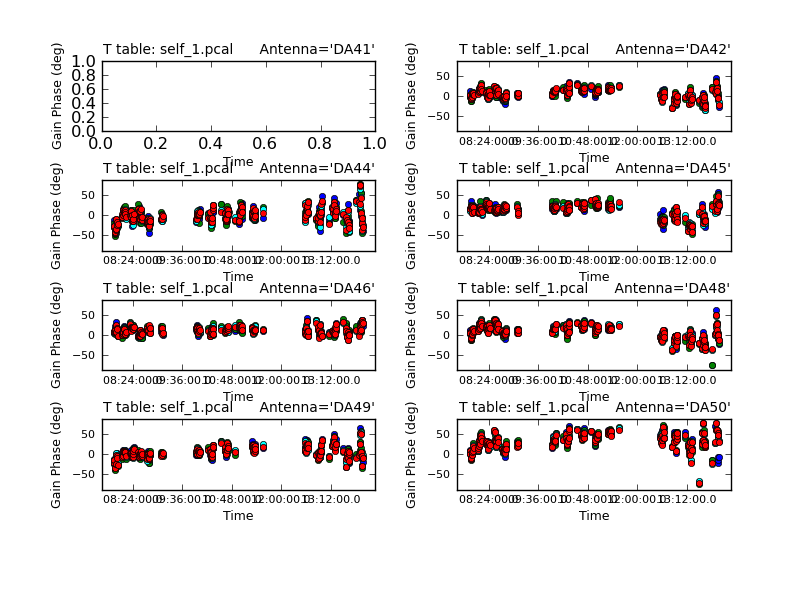
Next we'll examine the solutions using plotcal.
# In CASA
plotcal(caltable='self_1.pcal',xaxis='time',yaxis='phase',
spw='',field='',iteration='antenna',
subplot=421,plotrange=[0,0,-80,80],figfile='self_1_phase.png')
The magnitude of corrections gives you a sense of how accurate the phase transfer from the calibrators was. You are checking to see that the phases are being well tracked by the solutions, i.e. smoothly varying functions of time within a given scan. Also the solutions for each spectral window should be nearly identical, indicating that the phase offsets between spectral windows are well calibrated already. Overall, these solutions look fine. If they didn't you could try increasing the solint and/or combining the spw into a single solution.
After checking that you are happy with the solutions, apply them with applycal
# In CASA
applycal(vis='TWHya_cont.ms',field='',gaintable=['self_1.pcal'],calwt=F)
<figure id="Viewer_pcal1.png">
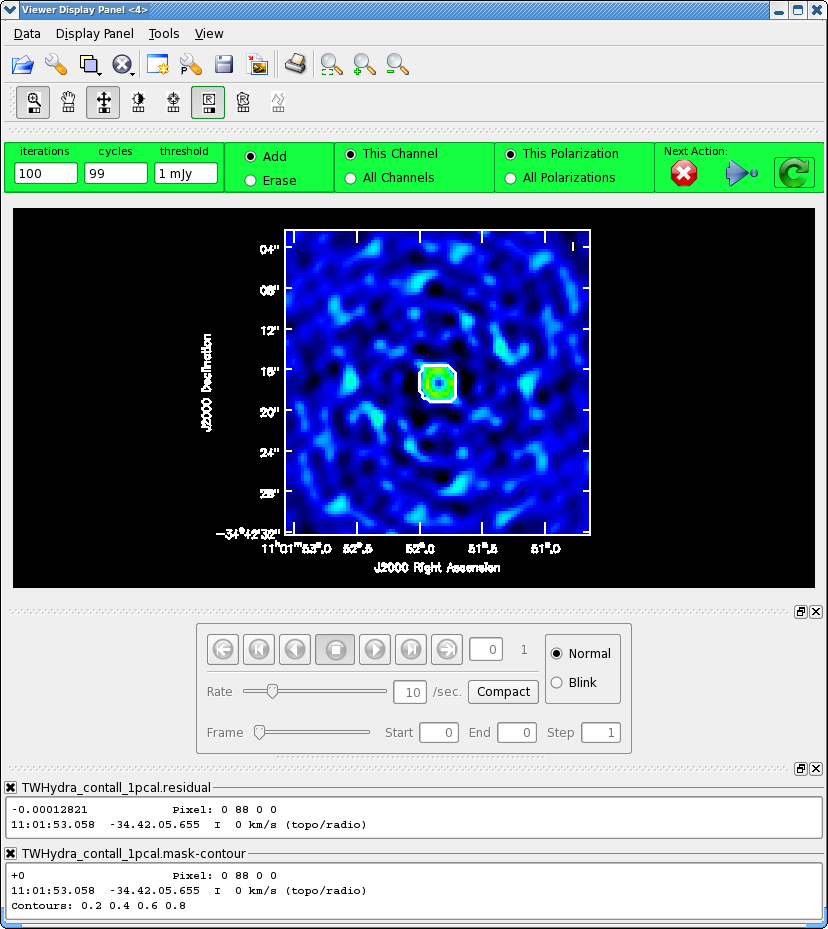
</figure>
Next clean the source again, to save time we can start with the clean mask from the last iteration. If you make interactive adjustments to the clean mask, they will be written to the new mask TWHydra.continuum.1pcal.mask, and not the old one.
# In CASA
os.system('rm -rf TWHya.continuum.1pcal.*')
clean(vis='TWHya_cont.ms',imagename='TWHya.continuum.1pcal',
mode='mfs',imagermode='csclean',
imsize=300,cell=['0.1arcsec'],spw='',
weighting='briggs',robust=0.5,
mask='TWHya.continuum.mask',usescratch=False,
interactive=T,threshold='1mJy',niter=10000)
Now after the first 500 iterations you see that the residuals inside the clean mask are higher than last time. This is because flux that had been spread out in the map due to poorly correlated phases has been placed where it belongs on the source, thereby increasing its flux. Let's continue cleaning for 1200 iterations.
Now let's compare the maps made before and after self-calibration using imview.
# In CASA
imview(raster=[{'file':'TWHya.continuum.image','range':[-0.01,0.05]},
{'file':'TWHya.continuum.1pcal.image','range':[-0.01,0.05]}])
Once the Viewer Display panel opens, click on the box next in the "Images" panel and then use the tapedeck buttons in the panel to alternate between the two images. We use the range parameter to put the two images on the same color scale. It is obvious that the self-calibrated image has much better residuals. To quantify the improvement in rms noise, draw a region that excludes the central source with the rectangle tool and then double click inside. Repeat that process for the second image. Statistics for the region will print to the terminal.:
(TWHya.continuum.image)
Stokes Velocity Frame Doppler Frequency
I -10.5133km/s LSRK RADIO 3.65287e+11
BrightnessUnit BeamArea Npts Sum FluxDensity
Jy/beam 25.4321 20100 -8.748308e-01 -3.439866e-02
Mean Rms Std dev Minimum Maximum
-4.352392e-05 2.133304e-03 2.132913e-03 -7.120122e-03 5.782832e-03
region count
1
(TWHya.continuum.1pcal.image)
Stokes Velocity Frame Doppler Frequency
I -10.5133km/s LSRK RADIO 3.65287e+11
BrightnessUnit BeamArea Npts Sum FluxDensity
Jy/beam 25.4701 20100 -4.660212e-02 -1.829681e-03
Mean Rms Std dev Minimum Maximum
-2.318513e-06 4.291876e-04 4.291920e-04 -1.593901e-03 1.666186e-03
region count
1
The self-calibrated image has improved the rms noise by a factor of 5.
Second cycle of self-calibration: phase only
Now that we have a better model, we can try to determine phase solutions again using a second round of phase-only self calibration. We will write the gain solutions to a new table, but these solutions will replace the solutions we determined in the first round. It may be useful to experiment with the solution interval. Keep in mind that the integration time is 6.05 seconds for this observation. (The integration time we refer to here is the "dump time" of the correlator, not the cumulative on-source time.)
In this example, we will stick with the 30 second solution interval.
# In CASA
os.system('rm -rf self_2*')
gaincal(vis='TWHya_cont.ms',caltable='self_2.pcal',
solint='30s',combine='',gaintype='T',
refant='DV22',spw='',minblperant=4,
calmode='p',minsnr=2)
Now plot the new solutions.
# In CASA
plotcal(caltable='self_2.pcal',xaxis='time',yaxis='phase',
spw='',field='',iteration='antenna',
subplot=421,plotrange=[0,0,-80,80],figfile='self_2_phase.png')
The solutions look much the same as those after the first round of phase calibration, as expected, since the corrections from the (slightly better) new model are minor in this case. We can apply the new phase solutions and make a new image to see if there is improvement.
# In CASA
applycal(vis='TWHya_cont.ms',field='',gaintable=['self_2.pcal'],calwt=F)
Note, here, that applycal applies the gain solutions to the raw data and overwrites any results in the "corrected data" column of the measurement set, so this calibration replaces what we applied after the first-round solutions.
Now clean again, updating mask with the most recent mask file.
# In CASA
os.system('rm -rf TWHya.continuum.2pcal.*')
clean(vis='TWHya_cont.ms',imagename='TWHya.continuum.2pcal',
mode='mfs',imagermode='csclean',
imsize=300,cell=['0.1arcsec'],spw='',
weighting='briggs',robust=0.5,usescratch=False,
mask='TWHya.continuum.1pcal.mask',
interactive=T,threshold='1mJy',niter=10000)
Clean again, interactively. In general, you should pay attention to the total flux being cleaned after each round, and the residuals inside the clean box as compared to those outside. When the cleaned flux is small and the residuals match, the image has been sufficiently cleaned. Stop by hitting the red X on the GUI. In this case, let's again clean for 1200 iterations. The task reports a total cleaned flux and residuals similar to what we saw after the first round of cleaning:
Model 0: max, min residuals = 0.00105882, -0.00017499 clean flux 1.78282
Now repeat the display, blinking, and image statistics steps described above for the 1st and 2nd phase-only self-cal iterations.
# In CASA
imview(raster=[{'file':'TWHya.continuum.1pcal.image','range':[-0.01,0.05]},
{'file':'TWHya.continuum.2pcal.image','range':[-0.01,0.05]}])
The second round made an improvement, but the difference is small:
(TWHya.continuum.1pcal.image)
Stokes Velocity Frame Doppler Frequency
I -10.5133km/s LSRK RADIO 3.65287e+11
BrightnessUnit BeamArea Npts Sum FluxDensity
Jy/beam 25.4696 21576 -7.237051e-02 -2.841450e-03
Mean Rms Std dev Minimum Maximum
-3.354213e-06 4.405892e-04 4.405866e-04 -1.534719e-03 1.791427e-03
region count
1
---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ----
(TWHya.continuum.2pcal.image)
Stokes Velocity Frame Doppler Frequency
I -10.5133km/s LSRK RADIO 3.65287e+11
BrightnessUnit BeamArea Npts Sum FluxDensity
Jy/beam 25.3159 21576 -6.708313e-02 -2.649847e-03
Mean Rms Std dev Minimum Maximum
-3.109155e-06 4.225414e-04 4.225398e-04 -1.471975e-03 1.751540e-03
region count
1
This small improvement suggests that we have gone as far as we can with phase self-cal.
Third cycle of self-calibration: amplitude & phase
Next we will add amplitude self-calibration to the process. Amplitude changes more slowly than phase, so we will determine one solution per scan. We can set solint = 'inf', which tells gaincal to use the "combine" parameter to determine how to get the solution interval. With combine set to the default (combine = ) we will get one solution per scan.
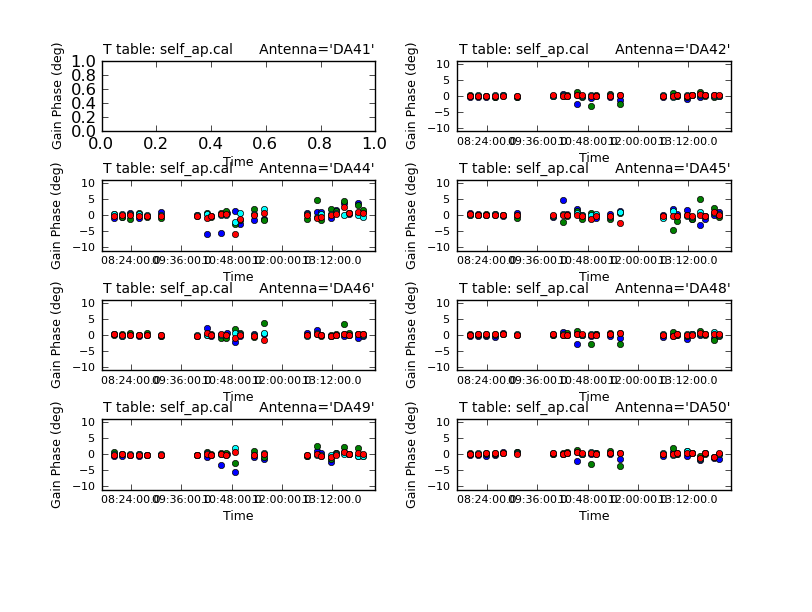
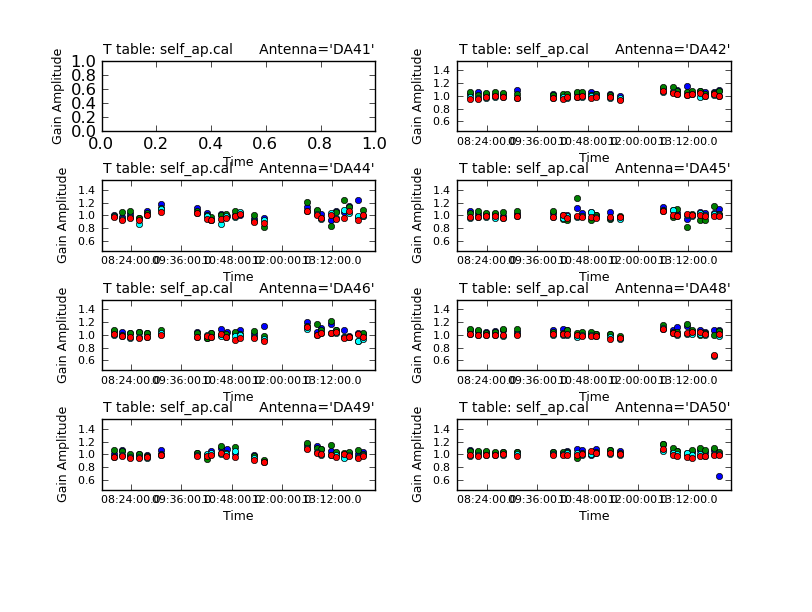
Also, note that we set gaintable to 'self_2.pcal'. This tells gaincal to apply the former phase-only solutions prior to determining the new amplitude+phase solutions. So, the new solutions will be incremental. Later when we apply these solutions to the data sets, we must be sure to apply both the phase-only solutions and the amplitude+phase solutions.
<figure id="C0Guide_self_ap_phase.png">
</figure>
<figure id="C0Guide_self_ap_amp.png">
</figure>
# In CASA
gaincal(vis='TWHya_cont.ms',caltable='self_ap.cal',
solint='inf',combine='',gaintype='T',
refant='DV22',spw='',minblperant=4,
gaintable=['self_2.pcal'],
calmode='ap',minsnr=2)
plotcal(caltable='self_ap.cal',xaxis='time',yaxis='phase',
spw='',field='',antenna='',iteration='antenna',
subplot=421,plotrange=[0,0,-10,10],figfile='self_ap_phase.png')
plotcal(caltable='self_ap.cal',xaxis='time',yaxis='amp',
spw='',field='',antenna='',iteration='antenna',
subplot=421,plotrange=[0,0,0.5,1.5],figfile='self_ap_amp.png')
Use the "next" button on the plotcal GUI to advance the plot to see additional antennas. Since phase corrections have already been applied, we see that the new incremental phase solutions are fairly small. The amplitude corrections are at the level of ~10% in many cases, but there is considerable variance among different antennas.
We can apply the calibration now with applycal. Note we apply two gain tables.
# In CASA
applycal(vis='TWHya_cont.ms',field='',gaintable=['self_2.pcal','self_ap.cal'],calwt=F)
<figure id="C0Guide_selfcal_time.png">
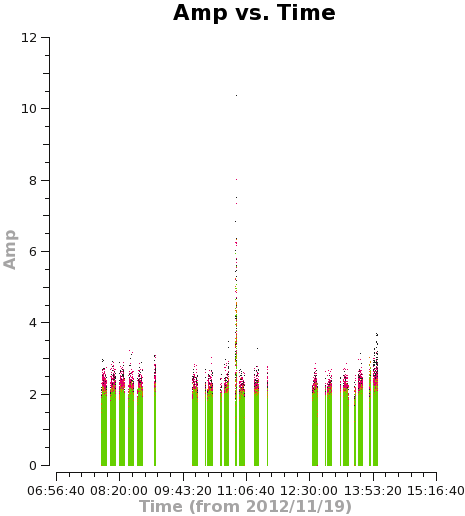
</figure>
<figure id="C0Guide_selfcal_uvdist.png">
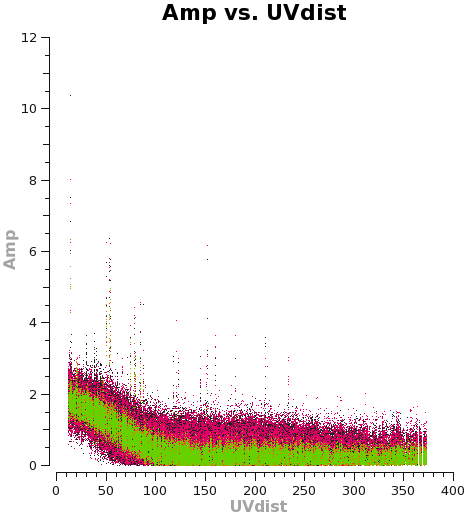
</figure>
Let's plot the u,v data against both time and u,v-distance. It is a good idea to check these after an amplitude self-calbration to be sure that it worked properly. Occasionally, a few spurious bits of data will get blown up by the amplitude self-cal.
# In CASA
plotms(vis='TWHya_cont.ms',xaxis='time',yaxis='amp',
avgchannel='38',ydatacolumn='corrected',coloraxis='spw',
plotfile='selfcal_time.png')
# In CASA
plotms(vis='TWHya_cont.ms',xaxis='uvdist',yaxis='amp',
avgchannel='38',ydatacolumn='corrected',coloraxis='spw',
plotfile='selfcal_uvdist.png')
Using the "locate" tool in the plotms GUI, select a few bad data points and confirm the scan number. We can see that bas data points exist for, at least, spw 0 and 1. To be conservative, we can flag all of scan 72. It's not much data compared to the full observation.
# In CASA
flagdata(vis='TWHya_cont.ms', mode='manual', scan='72')
Replot the u,v data to confirm the bad data have been removed. IMPORTANT: when re-plotting this data set in plotms, you must remember to select the "force reload" button on the GUI. Otherwise, the plot will not update with the new flags.
If the plots not look good, you can proceed to make the final continuum image.
# In CASA
os.system('rm -rf TWHya.continuum.apcal.*')
clean(vis='TWHya_cont.ms',imagename='TWHya.continuum.apcal',
mode='mfs',imagermode='csclean',
imsize=300,cell=['0.1arcsec'],spw='',
weighting='briggs',robust=0.5,usescratch=False,
interactive=T,threshold='0.1mJy',niter=5000,npercycle=1000)
<figure id="C0Guide_cont_clean_residuals.png">
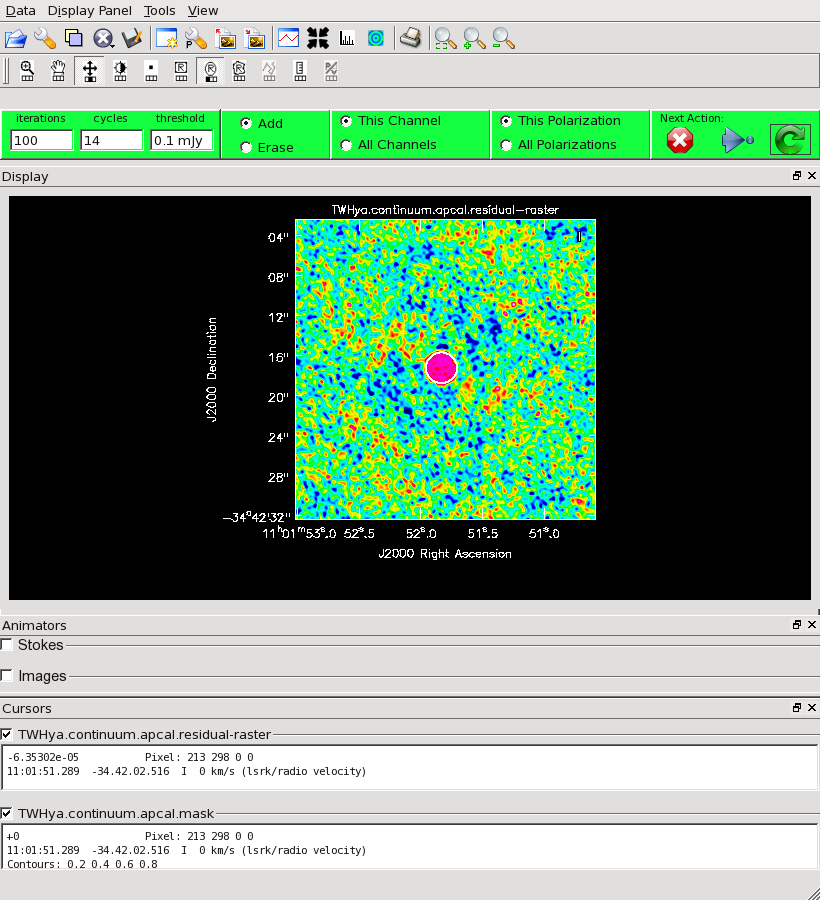
</figure>
You will need to execute over a thousand iterations in total to get the image fully cleaned, so we start the clean process with 1000 iterations and press the green circle. In the GUI, you should then adjust the number of iterations per cycle down to 100 for additional cleaning. Update the clean box if necessary, by expanding the region with a larger ellipse. Note that the pattern of residuals inside the clean box may not come to match that outside it, but you want to clean until the magnitude of the residuals is similar, inside it and out. In this case, the cleaning takes about 1500 iterations.
The clean task reports a total cleaned flux of 1.76 Jy.
# In CASA
imview(raster=[{'file':'TWHya.continuum.2pcal.image','range':[-0.01,0.05]},
{'file':'TWHya.continuum.apcal.image','range':[-0.01,0.05]}])
(TWHya.continuum.2pcal.image)
Stokes Velocity Frame Doppler Frequency
I -10.5133km/s LSRK RADIO 3.65287e+11
BrightnessUnit BeamArea Npts Sum FluxDensity
Jy/beam 25.3159 20553 1.654666e-01 6.536086e-03
Mean Rms Std dev Minimum Maximum
8.050727e-06 4.115417e-04 4.114729e-04 -1.471975e-03 1.612787e-03
region count
1
---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ----
(TWHya.continuum.apcal.image)
Stokes Velocity Frame Doppler Frequency
I -10.5133km/s LSRK RADIO 3.65287e+11
BrightnessUnit BeamArea Npts Sum FluxDensity
Jy/beam 25.3653 20553 3.826559e-02 1.508581e-03
Mean Rms Std dev Minimum Maximum
1.861801e-06 2.410208e-04 2.410195e-04 -1.023332e-03 8.470871e-04
region count
1
We see that the amplitude+phase self-cal made a significant improvement in the rms of the image, compared to the phase-only self-cal from 0.41 mJy to 0.24 mJy.
Apply Calibration to the Full Data Set and Split the Line Data
Now we apply the self-calibration derived from the continuum emission to the full-resolution data.
# In CASA
applycal(vis='calibrated.ms',gaintable=['self_2.pcal','self_ap.cal'],calwt=F)
Remember to flag the bad scan, 72.
# In CASA
flagdata(vis='calibrated.ms',scan='72')
Next we can split off the spectral line data for further processing.
# In CASA
os.system('rm -rf TWHydra_H2D+.ms*')
split(vis='calibrated.ms',outputvis='TWHydra_H2D+.ms',datacolumn='corrected',spw='0',field='TW Hya')
os.system('rm -rf TWHydra_N2H+.ms*')
split(vis='calibrated.ms',outputvis='TWHydra_N2H+.ms',datacolumn='corrected',spw='1',field='TW Hya')
Subtracting the Continuum
The next step in the processing is to subtract the continuum from the full data set, in preparation for imaging the line emission. We'll do the subtraction in the u,v plane. Usually, we would determine the continuum from line-free channels, so we begin by plotting the u,v amplitudes using plotms.
# In CASA
plotms(vis='TWHydra_N2H+.ms',xaxis='channel',yaxis='amp',avgtime='1e8',avgscan=T)
In this data set, the line emission is evident directly in the u,v amplitudes. The line is not strong enough to impact continuum subtraction, but during continuum subtraction we will avoid the channels with line emission in any case. In fitting for the continuum we will also avoid a few channels at the band edges, where there can sometimes be erratic data.
# In CASA
uvcontsub(vis='TWHydra_N2H+.ms',fitorder=1,fitspw='0:20~2400,0:2500~3800')
Imaging N2H+
The final step is to clean the image cube. In this instance, we will follow the guideline in the scriptForImaging.py python script that is included in the data package, and image a subset of spectral channels near the expected N2H+ line. We will image in "velocity" mode, so clean will bin the u,v data according to the velocity specifications we provide: 20 channels spaced by 0.5 km/s.
# In CASA
os.system('rm -rf line.N2H+.*')
clean(vis = 'TWHydra_N2H+.ms.contsub', imagename = 'line.N2H+', mode = 'velocity', nchan = 20, start = '-2.5km/s',
width = '0.5km/s', outframe = 'LSRK', restfreq = '372.6725GHz', interpolation = 'nearest', imagermode = 'mosaic',
interactive=T, niter=500, imsize = [300, 300], cell = '0.1arcsec', weighting = 'briggs', robust = 0.5)