Baseband Stitching: Difference between revisions
Created page with "Category:CASA == Overview == In order to maintain constant rms noise across an observed frequency range, one may "overlap" EVLA basebands, offsetting one by 0.5 times the s..." |
|||
| (15 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
In order to maintain constant rms noise across an observed frequency range, one may "overlap" EVLA basebands, offsetting one by 0.5 times the subband width. This way, the decreased sensitivity at subband edges can be edited out, and these frequency ranges replaced with data from the other baseband. | In order to maintain constant rms noise across an observed frequency range, one may "overlap" EVLA basebands, offsetting one by 0.5 times the subband width. This way, the decreased sensitivity at subband edges can be edited out, and these frequency ranges replaced with data from the other baseband. | ||
The contributed task <tt>stitch</tt> can be used to make this process simpler. Given a basic set of inputs, it will select channel ranges for each spectral window, which can then be used to run {split} to create a new measurement set (MS) (<tt>stitch</tt> can also run {split}, further simplifying things). | The contributed task <tt>stitch</tt> can be used to make this process simpler. Given a basic set of inputs, it will select channel ranges for each spectral window, which can then be used to run {{split}} to create a new measurement set (MS) (<tt>stitch</tt> can also run {{split}}, further simplifying things). | ||
Here, we demonstrate the use of the <tt>stitch</tt> task. For information on reducing the data after running <tt>stitch</tt>, see the other CASA Guides EVLA tutorials — the data can be treated in the standard way. | Here, we demonstrate the use of the <tt>stitch</tt> task. For information on reducing the data after running <tt>stitch</tt>, see the other CASA Guides EVLA tutorials — the data can be treated in the standard way. | ||
| Line 14: | Line 14: | ||
[[File:Task_stitch.py]] | [[File:Task_stitch.py]] | ||
[[File:Stitch.xml]] | [[File:Stitch.xml]] | ||
| Line 26: | Line 27: | ||
<source lang="bash"> | <source lang="bash"> | ||
# In the shell | |||
buildmytasks | buildmytasks | ||
</source> | </source> | ||
Once this completes, import the task into CASA: | |||
<source lang="python"> | |||
# In CASA | |||
execfile('mytasks.py') | |||
</source> | |||
You should now be able to look at the help and parameter listings of <tt>stitch</tt>: | |||
<source lang="python"> | |||
# In CASA | |||
inp stitch | |||
</source> | |||
<pre> | |||
# stitch :: Stitches together desired pieces of overlapping basebands. | |||
vis = '' # Name of input measurement set | |||
baseband1 = [] # List of first and last spectral windows in lower-frequency baseband (e.g., | |||
# [2,9]) | |||
baseband2 = [] # List of first and last spectral windows in higher-frequency baseband (e.g., | |||
# [10,17]) | |||
startchan = 10 # Starting channel number for lower baseband spectral windows | |||
numchan = 40 # Number of channels to use in lower baseband spectral windows | |||
dosplit = True # Split out selected data to a new MS? | |||
outputvis = '' # Name of output measurement set | |||
datacolumn = 'corrected' # Which data column(s) to split out | |||
field = '' # Select field using ID(s) or name(s) | |||
correlation = '' # Select correlations | |||
keepflags = False # If practical, keep *completely flagged rows* instead of dropping them. | |||
async = False # If true the taskname must be started using stitch(...) | |||
</pre> | |||
and | |||
<source lang="python"> | |||
# In CASA | |||
help stitch | |||
</source> | |||
== Inspecting data and choosing channel range == | |||
For the purposes of this guide, we are using EVLA test data that was taken in the overlapping subband mode. This dataset may be found in the EVLA Data Archive; the archive file id is <tt>TVER0002_sb2557689_1.55517.018916574074</tt>. Here, we have imported it to an MS called <tt>overlapSubband.ms</tt>. | |||
To start with, we have a look at the data with {{listobs}}: | |||
<source lang="python"> | |||
# In CASA: | |||
listobs('overlapSubband.ms') | |||
</source> | |||
The most important part of the output is the spectral window information: | |||
<pre> | |||
Spectral Windows: (18 unique spectral windows and 1 unique polarization setups) | |||
SpwID #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) Corrs | |||
0 64 TOPO 4736 2000 128000 RR RL LR LL | |||
1 64 TOPO 7836 2000 128000 RR RL LR LL | |||
2 64 TOPO 4488 2000 128000 RR RL LR LL | |||
3 64 TOPO 4616 2000 128000 RR RL LR LL | |||
4 64 TOPO 4744 2000 128000 RR RL LR LL | |||
5 64 TOPO 4872 2000 128000 RR RL LR LL | |||
6 64 TOPO 5000 2000 128000 RR RL LR LL | |||
7 64 TOPO 5128 2000 128000 RR RL LR LL | |||
8 64 TOPO 5256 2000 128000 RR RL LR LL | |||
9 64 TOPO 5384 2000 128000 RR RL LR LL | |||
10 64 TOPO 4552 2000 128000 RR RL LR LL | |||
11 64 TOPO 4680 2000 128000 RR RL LR LL | |||
12 64 TOPO 4808 2000 128000 RR RL LR LL | |||
13 64 TOPO 4936 2000 128000 RR RL LR LL | |||
14 64 TOPO 5064 2000 128000 RR RL LR LL | |||
15 64 TOPO 5192 2000 128000 RR RL LR LL | |||
16 64 TOPO 5320 2000 128000 RR RL LR LL | |||
17 64 TOPO 5448 2000 128000 RR RL LR LL | |||
</pre> | |||
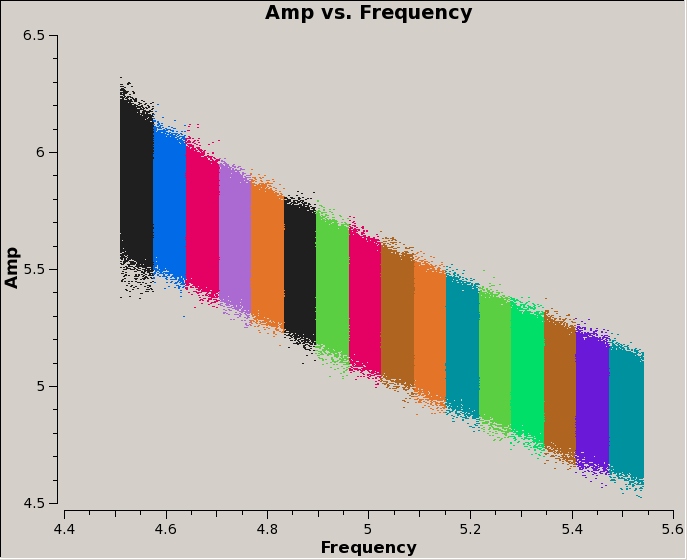
For this observation, we can see that spectral windows 0 and 1 are used for the "dummy" scans, then science data were taken in SPW 2-17. Furthermore, SPW 2-9 are contiguous and in the first baseband, then the frequency jumps back down again, and SPW 10-17 (second baseband) are shifted by half a SPW up in frequency relative to the first baseband. We can also see this by plotting the data: | |||
<source lang="python"> | |||
# In CASA: | |||
plotms(vis='overlapSubband.ms', field='1', antenna='ea21', xaxis='freq', | |||
yaxis='amp', coloraxis='spw', correlation='rr', avgtime='30s') | |||
</source> | |||
[[Image:overlapSB1.png|200px|thumb|right|overlapping subband observation]] | |||
Note that we're only plotting data for one field, antenna, and correlation, and averaging in time by 30 seconds. Also, we're coloring the data points by their spectral window, to show visually the overlap in frequency between the two basebands. | |||
Since the data have not yet been calibrated, the individual antenna variations are still apparent, as expected. Still, it is clear where the regions of reduced sensitivity lie: the amplitude drops off significantly at the edge of the very first and last spectral window in each baseband, as well as at the spectral window boundaries. | |||
== Running <tt>stitch</tt> == | |||
In order to run <tt>stitch</tt>, we must first determine which channels we would like to keep from the lower-frequency baseband. (Note that it will retain all the channels in the first SPW of the lower-frequency baseband, and all channels in the last SPW of the higher-frequency baseband, so that we don't lose more bandwidth than needed.) | |||
[[Image:overlapSB2.png|200px|thumb|right|overlapping subband zoom]] | |||
Specifically, we will need to determine the start channel and number of channels to keep. The main constraint will be that we want to avoid using the roll-off region in the last spectral window of the lower-frequency baseband. | |||
Going back to {{plotms}}, we first use the Zoom [[Image:ZoomButton.png]] button to look at this part of the data, then click on the Mark Regions [[Image:MarkRegionsButton.png]] button to locate a channel at the higher end of the spectral window (see plot). Finally, use the Locate [[Image:LocateButton.png]] button to determine the channel number. The output will be displayed in the logger: in this particular case, we see that it's channel 43 of SPW 9: | |||
<pre> | |||
Scan=19 Field=0137+331=3C48[1] Time=2010/11/17/00:53:55.9 BL=ea07@E14 & ea21@E12[6&20] Spw=9 Chan=43 Freq=5.48 Corr=RR X=5.48 Y=0.0949401 (557872/335/432) | |||
</pre> | |||
Now, we need to determine the starting channel. For this, we want to select a channel a few higher than the edge of the amplitude decrease; channel 11 seems like a good choice. Therefore, our selection for the lower-frequency baseband will be 33 channels, starting at channel 11. | |||
There are two ways to run the <tt>stitch</tt> task: you can either have it simply print out the SPW selection string to the logger and terminal, in which case you would run split later as a separate step, or you can ask it to split the data as well. | |||
=== Example 1: printing the SPW string only === | |||
As input, <tt>stitch</tt> needs to know the spectral window indices of the lower-frequency and higher-frequency basebands (baseband1 and baseband2), as well as the start channel and number of channels to use for baseband1. For our example, this would be: | |||
<source lang="python"> | |||
# In CASA: | |||
stitch(vis='overlapSubband.ms', baseband1=[2,9], baseband2=[10,17], \ | |||
startchan=11, numchan=33, dosplit=False) | |||
</source> | |||
This will print: | |||
<pre> | |||
Out[12]: '2:0~43,3~9:11~43,10~16:12~42,17:12~63' | |||
</pre> | |||
=== Example 2: determining the SPW string and splitting === | |||
We can also ask <tt>stitch</tt> to split out the desired data using the derived SPW string. In this case, we would input: | |||
<source lang="python"> | |||
# In CASA: | |||
stitch(vis='overlapSubband.ms', baseband1=[2,9], baseband2=[10,17], \ | |||
startchan=11, numchan=33, dosplit=True, | |||
outputvis='overlapSubband.stitch.ms', datacolumn='data') | |||
</source> | |||
If you have already calibrated the data before running <tt>stitch</tt>, you could use the 'corrected' data column here; you can also choose to keep particular polarizations and excise flagged data, if desired (see the <tt>stitch</tt> help file for full explanations of the parameters). | |||
[[Image:overlapSB3.png|200px|thumb|right|overlapping subband stitched]] | |||
Now, we can plot the stitched MS: | |||
<source lang="python"> | |||
# In CASA: | |||
plotms(vis='overlapSubband.stitch.ms', field='1', antenna='ea21', xaxis='freq', | |||
yaxis='amp', coloraxis='spw', correlation='rr', avgtime='30s') | |||
</source> | |||
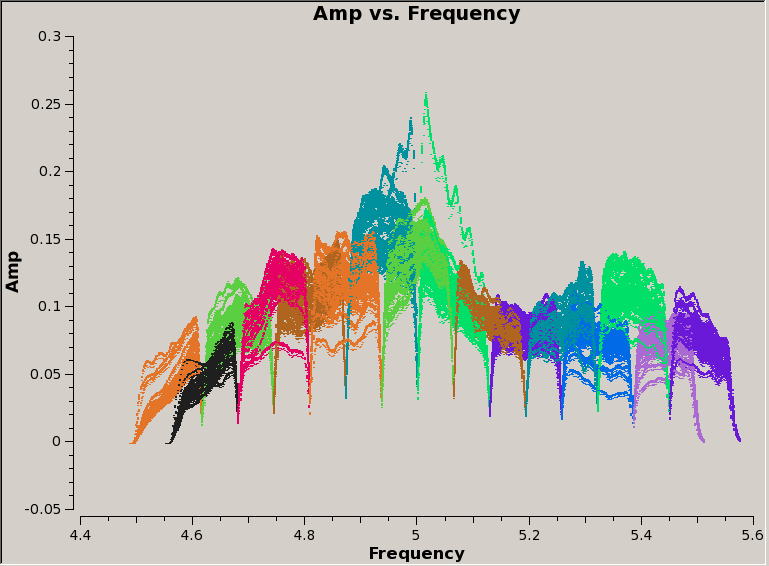
Notice that there are no longer any deep troughs in the amplitude, so we have successfully excised data which would have higher rms values. There are still mismatches between the spectral windows, but these will disappear when the data are calibrated. | |||
== Closing remarks == | |||
While we have chosen to run <tt>stitch</tt> before calibration, it would be perfectly fine to calibrate first as well. However, you will operating on more data than you need to in this case, so stitching first is optimal. | |||
You can calibrate the stitched MS as you would any other dataset — see the [[EVLA Tutorials]] page for examples of calibration and analysis. | |||
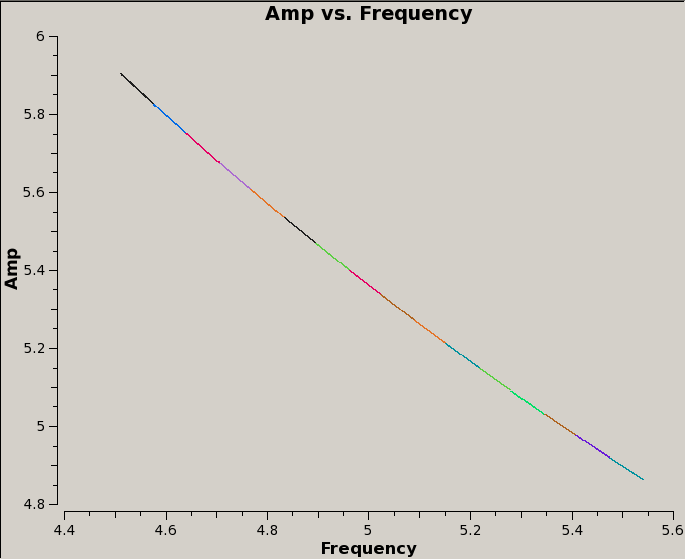
For the data presented here, standard flagging and calibration were done after stitching, and the corrected data for 3C48 are shown below. The first plot shows result of issuing the same {{plotms}} command as we did earlier; the second shows the result of averaging the data across antennas and over the time range of the observation. | |||
[[Image:overlapSB4.png|200px|thumb|left|overlapping subband calibrated]] | |||
[[Image:overlapSB5.png|200px|thumb|right|overlapping subband averaged]] | |||
Latest revision as of 01:09, 2 March 2012
Overview
In order to maintain constant rms noise across an observed frequency range, one may "overlap" EVLA basebands, offsetting one by 0.5 times the subband width. This way, the decreased sensitivity at subband edges can be edited out, and these frequency ranges replaced with data from the other baseband.
The contributed task stitch can be used to make this process simpler. Given a basic set of inputs, it will select channel ranges for each spectral window, which can then be used to run split to create a new measurement set (MS) (stitch can also run split, further simplifying things).
Here, we demonstrate the use of the stitch task. For information on reducing the data after running stitch, see the other CASA Guides EVLA tutorials — the data can be treated in the standard way.
Obtaining and installing stitch
First, you will need to download two files that are needed to build the task:
Because of a quirk in the way Wiki stores files, you need to rename these to no longer have capitalized first letters, task_stitch.py and stitch.xml. Next, run buildmytasks to build the needed files for the task. This can either be done in CASA, or in a UNIX shell. You must be inside the directory where you have saved the files.
# In CASA
!buildmytasks
or:
# In the shell
buildmytasks
Once this completes, import the task into CASA:
# In CASA
execfile('mytasks.py')
You should now be able to look at the help and parameter listings of stitch:
# In CASA
inp stitch
# stitch :: Stitches together desired pieces of overlapping basebands.
vis = '' # Name of input measurement set
baseband1 = [] # List of first and last spectral windows in lower-frequency baseband (e.g.,
# [2,9])
baseband2 = [] # List of first and last spectral windows in higher-frequency baseband (e.g.,
# [10,17])
startchan = 10 # Starting channel number for lower baseband spectral windows
numchan = 40 # Number of channels to use in lower baseband spectral windows
dosplit = True # Split out selected data to a new MS?
outputvis = '' # Name of output measurement set
datacolumn = 'corrected' # Which data column(s) to split out
field = '' # Select field using ID(s) or name(s)
correlation = '' # Select correlations
keepflags = False # If practical, keep *completely flagged rows* instead of dropping them.
async = False # If true the taskname must be started using stitch(...)
and
# In CASA
help stitch
Inspecting data and choosing channel range
For the purposes of this guide, we are using EVLA test data that was taken in the overlapping subband mode. This dataset may be found in the EVLA Data Archive; the archive file id is TVER0002_sb2557689_1.55517.018916574074. Here, we have imported it to an MS called overlapSubband.ms.
To start with, we have a look at the data with listobs:
# In CASA:
listobs('overlapSubband.ms')
The most important part of the output is the spectral window information:
Spectral Windows: (18 unique spectral windows and 1 unique polarization setups) SpwID #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) Corrs 0 64 TOPO 4736 2000 128000 RR RL LR LL 1 64 TOPO 7836 2000 128000 RR RL LR LL 2 64 TOPO 4488 2000 128000 RR RL LR LL 3 64 TOPO 4616 2000 128000 RR RL LR LL 4 64 TOPO 4744 2000 128000 RR RL LR LL 5 64 TOPO 4872 2000 128000 RR RL LR LL 6 64 TOPO 5000 2000 128000 RR RL LR LL 7 64 TOPO 5128 2000 128000 RR RL LR LL 8 64 TOPO 5256 2000 128000 RR RL LR LL 9 64 TOPO 5384 2000 128000 RR RL LR LL 10 64 TOPO 4552 2000 128000 RR RL LR LL 11 64 TOPO 4680 2000 128000 RR RL LR LL 12 64 TOPO 4808 2000 128000 RR RL LR LL 13 64 TOPO 4936 2000 128000 RR RL LR LL 14 64 TOPO 5064 2000 128000 RR RL LR LL 15 64 TOPO 5192 2000 128000 RR RL LR LL 16 64 TOPO 5320 2000 128000 RR RL LR LL 17 64 TOPO 5448 2000 128000 RR RL LR LL
For this observation, we can see that spectral windows 0 and 1 are used for the "dummy" scans, then science data were taken in SPW 2-17. Furthermore, SPW 2-9 are contiguous and in the first baseband, then the frequency jumps back down again, and SPW 10-17 (second baseband) are shifted by half a SPW up in frequency relative to the first baseband. We can also see this by plotting the data:
# In CASA:
plotms(vis='overlapSubband.ms', field='1', antenna='ea21', xaxis='freq',
yaxis='amp', coloraxis='spw', correlation='rr', avgtime='30s')
Note that we're only plotting data for one field, antenna, and correlation, and averaging in time by 30 seconds. Also, we're coloring the data points by their spectral window, to show visually the overlap in frequency between the two basebands.
Since the data have not yet been calibrated, the individual antenna variations are still apparent, as expected. Still, it is clear where the regions of reduced sensitivity lie: the amplitude drops off significantly at the edge of the very first and last spectral window in each baseband, as well as at the spectral window boundaries.
Running stitch
In order to run stitch, we must first determine which channels we would like to keep from the lower-frequency baseband. (Note that it will retain all the channels in the first SPW of the lower-frequency baseband, and all channels in the last SPW of the higher-frequency baseband, so that we don't lose more bandwidth than needed.)
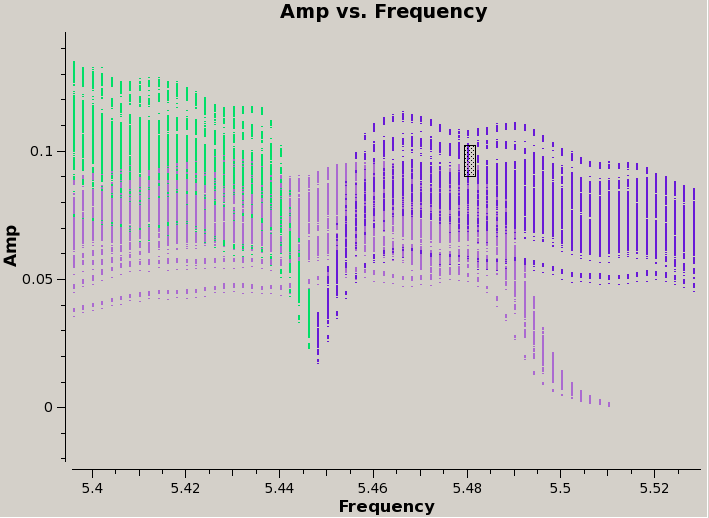
Specifically, we will need to determine the start channel and number of channels to keep. The main constraint will be that we want to avoid using the roll-off region in the last spectral window of the lower-frequency baseband.
Going back to plotms, we first use the Zoom ![]() button to look at this part of the data, then click on the Mark Regions
button to look at this part of the data, then click on the Mark Regions ![]() button to locate a channel at the higher end of the spectral window (see plot). Finally, use the Locate
button to locate a channel at the higher end of the spectral window (see plot). Finally, use the Locate ![]() button to determine the channel number. The output will be displayed in the logger: in this particular case, we see that it's channel 43 of SPW 9:
button to determine the channel number. The output will be displayed in the logger: in this particular case, we see that it's channel 43 of SPW 9:
Scan=19 Field=0137+331=3C48[1] Time=2010/11/17/00:53:55.9 BL=ea07@E14 & ea21@E12[6&20] Spw=9 Chan=43 Freq=5.48 Corr=RR X=5.48 Y=0.0949401 (557872/335/432)
Now, we need to determine the starting channel. For this, we want to select a channel a few higher than the edge of the amplitude decrease; channel 11 seems like a good choice. Therefore, our selection for the lower-frequency baseband will be 33 channels, starting at channel 11.
There are two ways to run the stitch task: you can either have it simply print out the SPW selection string to the logger and terminal, in which case you would run split later as a separate step, or you can ask it to split the data as well.
Example 1: printing the SPW string only
As input, stitch needs to know the spectral window indices of the lower-frequency and higher-frequency basebands (baseband1 and baseband2), as well as the start channel and number of channels to use for baseband1. For our example, this would be:
# In CASA:
stitch(vis='overlapSubband.ms', baseband1=[2,9], baseband2=[10,17], \
startchan=11, numchan=33, dosplit=False)
This will print:
Out[12]: '2:0~43,3~9:11~43,10~16:12~42,17:12~63'
Example 2: determining the SPW string and splitting
We can also ask stitch to split out the desired data using the derived SPW string. In this case, we would input:
# In CASA:
stitch(vis='overlapSubband.ms', baseband1=[2,9], baseband2=[10,17], \
startchan=11, numchan=33, dosplit=True,
outputvis='overlapSubband.stitch.ms', datacolumn='data')
If you have already calibrated the data before running stitch, you could use the 'corrected' data column here; you can also choose to keep particular polarizations and excise flagged data, if desired (see the stitch help file for full explanations of the parameters).
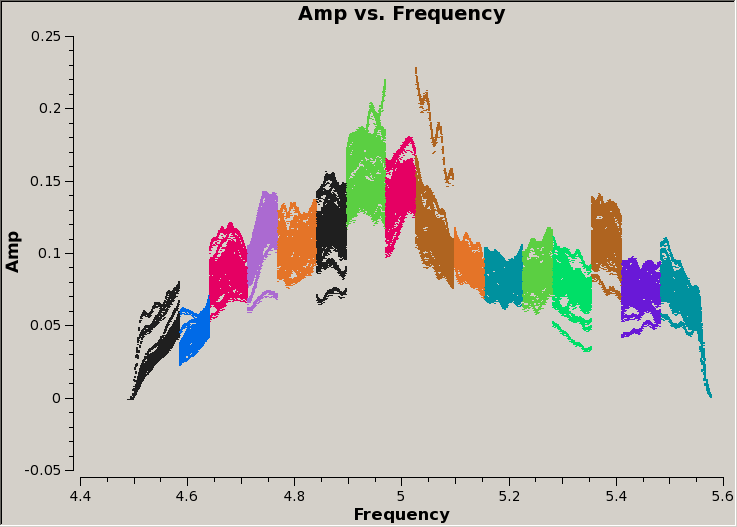
Now, we can plot the stitched MS:
# In CASA:
plotms(vis='overlapSubband.stitch.ms', field='1', antenna='ea21', xaxis='freq',
yaxis='amp', coloraxis='spw', correlation='rr', avgtime='30s')
Notice that there are no longer any deep troughs in the amplitude, so we have successfully excised data which would have higher rms values. There are still mismatches between the spectral windows, but these will disappear when the data are calibrated.
Closing remarks
While we have chosen to run stitch before calibration, it would be perfectly fine to calibrate first as well. However, you will operating on more data than you need to in this case, so stitching first is optimal.
You can calibrate the stitched MS as you would any other dataset — see the EVLA Tutorials page for examples of calibration and analysis.
For the data presented here, standard flagging and calibration were done after stitching, and the corrected data for 3C48 are shown below. The first plot shows result of issuing the same plotms command as we did earlier; the second shows the result of averaging the data across antennas and over the time range of the observation.