Antennae Band7 - Imaging for CASA 3.3: Difference between revisions
No edit summary |
|||
| (154 intermediate revisions by 6 users not shown) | |||
| Line 2: | Line 2: | ||
==Overview== | ==Overview== | ||
This section of the '''[[AntennaeBand7]]''' CASA Guide covers the imaging of the continuum and spectral line data. It begins where the '''[[Antennae Band7 - Calibration]]''' section left off. If you completed the Calibration section of the guide, then you | This section of the '''[[AntennaeBand7]]''' CASA Guide covers the imaging of the continuum and spectral line data. It begins where the '''[[Antennae Band7 - Calibration]]''' section left off. If you completed the Calibration section of the guide, then you already have the calibrated data sets. If you did not complete the Calibration section, then you can download the calibrated uv-data from the region closest to your location: | ||
[http://almascience.nrao.edu/almadata/sciver/ | [http://almascience.nrao.edu/almadata/sciver/AntennaeBand7 North America] | ||
[http://almascience.eso.org/almadata/sciver/ | [http://almascience.eso.org/almadata/sciver/AntennaeBand7 Europe] | ||
[http://almascience.nao.ac.jp/almadata/sciver/ | [http://almascience.nao.ac.jp/almadata/sciver/AntennaeBand7 East Asia] | ||
Then download the file 'Antennae_Band7_CalibratedData.tgz' to obtain the calibrated uvdata. | Then download the file 'Antennae_Band7_CalibratedData.tgz' to obtain the calibrated uvdata. | ||
Once the download has finished, unpack the file using 'tar -xvzf Antennae_Band7_CalibratedData.tgz' | Once the download has finished, unpack the file using | ||
You should have Antennae_South.cal.ms and Antennae_North.cal.ms in your working directory. Then start CASA with the 'casapy' command. Be sure you are using version | |||
> 'tar -xvzf Antennae_Band7_CalibratedData.tgz' | |||
once it's unpacked, change directory (cd) to Antennae_Band7_CalibratedData. You should have '''Antennae_South.cal.ms''' and '''Antennae_North.cal.ms''' in your working directory. Then start CASA with the 'casapy' command. Be sure you are using version casapy-stable r19407 or later. | |||
== Imaging Mosaics == | == Imaging Mosaics == | ||
Mosaics like other kinds of images are created in the CASA task {{clean}}. To invoke mosaic mode, you simply set the parameter '''imagermode='mosaic''''. The default subparameter '''ftmachine='mosaic'''' is then automatically set. This is a joint deconvolution algorithm that works in the uv-plane. A convolution of the primary beam patterns for each pointing in the | <pre style="background-color: #E0FFFF;"> | ||
If you are unfamiliar with the basic concepts of deconvolution and clean, pause here and | |||
review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/wilner_synthesis10.pdf | |||
</pre> | |||
Mosaics like other kinds of images are created in the CASA task {{clean}}. To invoke mosaic mode, you simply set the parameter '''imagermode='mosaic''''. The default subparameter '''ftmachine='mosaic'''' is then automatically set. This is a joint deconvolution algorithm that works in the uv-plane. A convolution of the primary beam patterns for each pointing in the mosaic is created: the primary beam response function. The corresponding image of the mosaic response function will be called <imagename>.flux.pbcoverage and <imagename>.flux (where the latter differs from the former only if the sensitivity of each field in the mosaic varies). | |||
Well-sampled mosaics like the patterns observed for the Northern and Southern Antennae mosaics shown on the [[AntennaeBand7#ALMA_Data_Overview | Overview]] page are well-suited to joint deconvolution. However, if you have a poorly-sampled, or very irregular mosaic you may need to use the slower '''ftmachine='ft'''' which combines data in the image plane, similar to what is done in other packages like miriad. | Well-sampled mosaics like the patterns observed for the Northern and Southern Antennae mosaics shown on the [[AntennaeBand7#ALMA_Data_Overview | Overview]] page are well-suited to joint deconvolution. However, if you have a poorly-sampled, or very irregular mosaic you may need to use the slower '''ftmachine='ft'''' which combines data in the image plane, similar to what is done in other packages like miriad. | ||
Additionally, for mosaics it is essential to pick the center of the region to be imaged explicitly using the '''phasecenter''' parameter. Otherwise it will default to the first pointing included in the '''field''' parameter -- since this is often at one corner of the mosaic, the image will not be centered. For the Northern mosaic, the center pointing corresponds to field id=12. Note that during the final '''split''' in the calibration section that selected only the Antennae fields, the field ids were renumbered, so that the original centers (shown in the [[AntennaeBand7#ALMA_Data_Overview | Overview]]) have changed: field id 14 | Additionally, for mosaics it is essential to pick the center of the region to be imaged explicitly using the '''phasecenter''' parameter. Otherwise it will default to the first pointing included in the '''field''' parameter -- since this is often at one corner of the mosaic, the image will not be centered. For the Northern mosaic, the center pointing corresponds to field id=12. Note that during the final '''split''' in the calibration section that selected only the Antennae fields, the field ids were renumbered, so that the original centers (shown in the [[AntennaeBand7#ALMA_Data_Overview | Overview]]) have changed: field id 14 becomes 12 for the Northern mosaic and field 18 becomes 15 for the Southern mosaic. You can also set an explicit coordinate (see the {{clean}} help for syntax). | ||
<pre style="background-color: #E0FFFF;"> | |||
If you want to learn more about mosaicing, pause here and | |||
review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/jott-mosaicking-school-04.pdf | |||
</pre> | |||
== Continuum Imaging == | == Continuum Imaging == | ||
[[File:Antennae_South-AMPvsCH.png|200px|thumb|right|'''Fig. 1.''' Southern mosaic: Amplitude vs. channel. The CO(3-2) line is seen from 50 to 100]] | |||
[[File:Antennae_North-AMPvsCH.png|200px|thumb|right|'''Fig. 2.''' Northern mosaic: Amplitude vs. channel. The CO(3-2) line is seen from 70 to 100]] | |||
We will make 345 GHz continuum images for the two regions covered by the mosaics. We use the task {{clean}} over the channels that are free of the line emission; we avoid the edge channels which tend to be noisier due to bandpass rolloff effects. The line-free channels are found by plotting the average spectrum (all fields). We find the CO(3-2) line from channels 50 to 100 in the southern mosaic (Figure 1), and from 70 to 100 in the northern mosaic (Figure 2). | |||
We will make 345 GHz continuum images for the two regions covered by the mosaics. We use the task {{clean}} over the channels that are free of the line emission; we avoid the edge channels which tend to be noisier due to bandpass rolloff effects. The line-free channels are found by plotting the average spectrum. We find the CO(3-2) line from channels 50 to 100 in the southern mosaic, and from 70 to 100 in the northern mosaic. | |||
<source lang="python"> | |||
# In CASA | |||
plotms(vis='Antennae_North.cal.ms',xaxis='channel',yaxis='amp', | |||
avgtime='1e8',avgscan=T,plotfile='Antennae_North-AMPvsCH.png') | |||
</source> | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='Antennae_South.cal.ms',xaxis='channel',yaxis='amp', | plotms(vis='Antennae_South.cal.ms',xaxis='channel',yaxis='amp', | ||
avgtime='1e8',avgscan=T | avgtime='1e8',avgscan=T,plotfile='Antennae_South-AMPvsCH.png') | ||
</source> | </source> | ||
| Line 41: | Line 56: | ||
==== Northern Continuum Mosaic ==== | ==== Northern Continuum Mosaic ==== | ||
For illustrative purposes we first make a dirty image to see if there is emission and what the exact beam size is. It should be on the order of 1" but this will vary a bit according to the uv-coverage in the actual data. We will start with a '''cell''' size of 0.2" to oversample the beam by a factor of 5. The imsize needs to be large enough given the cell size to comfortably encompass the mosaic. From the mosaic footprints shown in the [[AntennaeBand7#ALMA_Data_Overview | overview]] we can see that Northern mosaic '''imsize''' needs to be about 1 arcmin. | For illustrative purposes we first make a dirty image to see if there is emission and what the exact beam size is. It should be on the order of 1" but this will vary a bit according to the uv-coverage in the actual data. We will start with a '''cell''' size of 0.2" to oversample the beam by a factor of 5. The imsize needs to be large enough given the cell size to comfortably encompass the mosaic. From the mosaic footprints shown in the [[AntennaeBand7#ALMA_Data_Overview | overview]], we can see that the Northern mosaic '''imsize''' needs to be about 1 arcmin. With 0.2" pixels, this requires a '''imsize'''=300. | ||
<u>Other essential {{clean}} parameters for this case include:</u> | <u>Other essential {{clean}} parameters for this case include:</u> | ||
*'''vis'''='Antennae_North.cal.ms': The calibrated dataset on the science target | *'''vis'''='Antennae_North.cal.ms': The calibrated dataset on the science target. {{clean}} will always use the CORRECTED DATA column (if it exists). | ||
*'''imagename=''''Antennae_North.Cont. | *'''imagename=''''Antennae_North.Cont.Dirty': The base name of the output images: | ||
** <imagename>.image # the final restored image | ** <imagename>.image # the final restored image | ||
** <imagename>.flux # the effective primary beam response (e.g. for pbcor) | ** <imagename>.flux # the effective primary beam response (e.g. for pbcor) | ||
| Line 53: | Line 68: | ||
** <imagename>.psf # the synthesized (dirty) beam | ** <imagename>.psf # the synthesized (dirty) beam | ||
*'''spw'''='0:1~50;120~164': To specify only the line-free channels of spectral window 0. | *'''spw'''='0:1~50;120~164': To specify only the line-free channels of spectral window 0. | ||
*'''mode='mfs'''': Multi-Frequency Synthesis: The default mode, which produces one image from all the specified data combined. This will grid each channel independently before imaging. For wide | *'''mode='mfs'''': Multi-Frequency Synthesis: The default mode, which produces one image from all the specified data combined. This will grid each channel independently before imaging. For wide bandwidths this will give a far superior result compared to averaging the channels and then imaging. | ||
*'''restfreq'''='345.79599GHz': The rest frequency of CO(3-2) can be found with [http://www.splatalogue.net/ splatalogue]. | |||
*'''niter'''=0: Maximum number of clean iterations -- '''niter=0 will do no cleaning''' | *'''niter'''=0: Maximum number of clean iterations -- '''niter=0 will do no cleaning''' | ||
| Line 68: | Line 84: | ||
</source> | </source> | ||
The reported beam size is 1. | The reported beam size is about 1.27" x 0.71" P.A. 86.142 degrees. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 75: | Line 91: | ||
</source> | </source> | ||
[[Image:North_cont_clean.png|200px|thumb|right|Fig. | <pre style="background-color: #E0FFFF;"> | ||
If you are unfamiliar with the CASA viewer it is a good idea to pause here and watch the demo at | |||
http://casa.nrao.edu/CasaViewerDemo/casaViewerDemo.html | |||
</pre> | |||
[[Image:North_cont_clean.png|200px|thumb|right|'''Fig. 3a.''' Residual for Northern continuum mosaic after 100 iterations; the clean mask is shown by the white contour.]] | |||
Yes there is definitely a detection in the vicinity of the Northern nucleus (see | Yes there is definitely a detection in the vicinity of the Northern nucleus (see Figs. 4 and 5 below). Using the square region icon, and drawing a box near but not including the emission, we find the rms noise is about 0.4 mJy/beam in the dirty image. | ||
Next we switch to refined values of '''cell'''=0.13" and '''imsize'''=500 based on the observed beam size. We also switch to interactive mode so that you can create a clean mask using the polygon tool (note you need to double click inside the polygon region to activate the mask. See [[TWHydraBand7_Imaging#Image_and_Self-Calibrate_the_Continuum | | Next we switch to refined values of '''cell'''=0.13" and '''imsize'''=500 based on the observed beam size. We also switch to interactive mode so that you can create a clean mask using the polygon tool (note you need to double click inside the polygon region to activate the mask). See [[TWHydraBand7_Imaging#Image_and_Self-Calibrate_the_Continuum | TWHya casaguide]] for a more complete description of interactive clean and mask creation. | ||
*'''niter'''=1000: Maximum number of clean iterations -- '''we will stop interactively''' | *'''niter'''=1000: Maximum number of clean iterations -- '''we will stop interactively''' | ||
*'''threshold'''='0.4mJy': Stop cleaning if the maximum residual is below this value (the dirty rms noise) | *'''threshold'''='0.4mJy': Stop cleaning if the maximum residual is below this value (the dirty rms noise) | ||
*'''interactive'''=T: Clean will be periodically | *'''interactive'''=T: Clean will be periodically interrupted to show the residual clean image. Interactive clean mask can be made. If no mask is created, no cleaning is done. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 95: | Line 115: | ||
imagermode='mosaic', | imagermode='mosaic', | ||
imsize=500,cell='0.13arcsec', | imsize=500,cell='0.13arcsec', | ||
interactive=T | interactive=T, | ||
niter=1000, threshold='0.4mJy') | niter=1000, threshold='0.4mJy') | ||
</source> | </source> | ||
The residuals are "noise-like" after only | The residuals are "noise-like" after only 100 iterations (see Fig. 3a), so hit the red X symbol in the interactive window to stop cleaning here. | ||
Note that if you run the {{clean}} task again with the same '''imagename''', without deleting the existing <imagename>.* files, {{clean}} assumes that you want to continue cleaning the existing images. We put an rm command before the clean command to guard against this, but sometimes this is what you want. | Note that if you run the {{clean}} task again with the same '''imagename''', without deleting the existing <imagename>.* files, {{clean}} assumes that you want to continue cleaning the existing images. We put an rm command before the clean command to guard against this, but sometimes this is what you want. | ||
| Line 105: | Line 125: | ||
==== Southern Continuum Mosaic ==== | ==== Southern Continuum Mosaic ==== | ||
[[Image:South_cont_clean.png|200px|thumb|right|Fig. | [[Image:South_cont_clean.png|200px|thumb|right|'''Fig. 3b.''' Residual for Southern continuum mosaic after 100 iterations; the clean mask is shown by the white contour.]] | ||
For the southern mosaic we modify the | For the southern mosaic we modify the '''phasecenter''', mosaic '''imsize''', and the line-free channels ('''spw''') to be consistent for this mosaic. We also bypass the dirty image step we did above for the Northern mosaic. As above, the mosaic size can be judged from [[AntennaeBand7#ALMA_Data_Overview | here]]. We expect the beam to be about the same and use the same '''cell''' size. During {{clean}}, we repeat the process of making a clean mask. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 118: | Line 138: | ||
imagermode='mosaic', | imagermode='mosaic', | ||
imsize=750,cell='0.13arcsec', | imsize=750,cell='0.13arcsec', | ||
interactive=T | interactive=T, | ||
niter=1000, threshold='0.4mJy') | niter=1000, threshold='0.4mJy') | ||
</source> | </source> | ||
The beam size reported in the logger for the Southern mosaic: 1. | The beam size reported in the logger for the Southern mosaic: 1.11"x 0.71", and P.A.=65 deg; which is a little better than the beam for the North owing to better uv-coverage. | ||
Stop after | Stop after 100 iterations (Fig. 3b). | ||
==== Image Statistics ==== | ==== Image Statistics ==== | ||
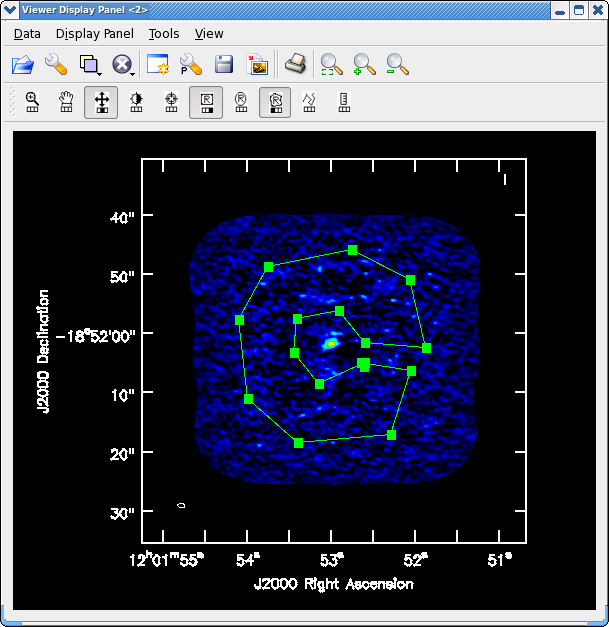
[[File:North_contstat.png|200px|thumb|right|Fig. 4. Example polygon for rms determination using the viewer.]] | [[File:North_contstat.png|200px|thumb|right|'''Fig. 4.''' Example polygon for rms determination using the viewer.]] | ||
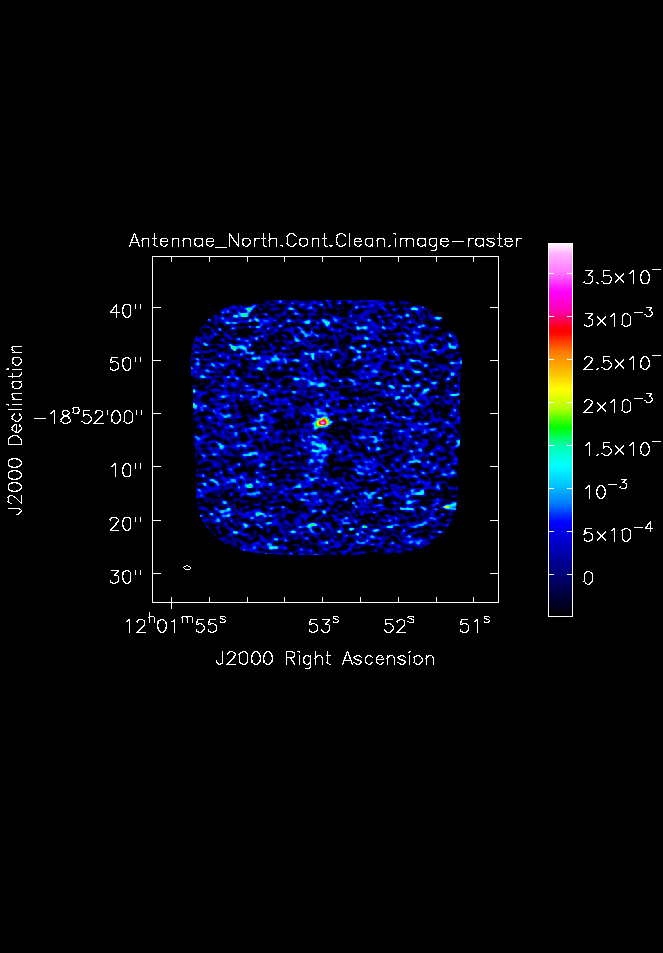
[[File:Antennae_North.Cont.Clean.image.png|200px|thumb|right|Fig. 5. 345 GHz continuum image of the northern mosaic made with imview.]] | [[File:Antennae_North.Cont.Clean.image.png|200px|thumb|right|'''Fig. 5.''' 345 GHz continuum image of the northern mosaic made with {{imview}}.]] | ||
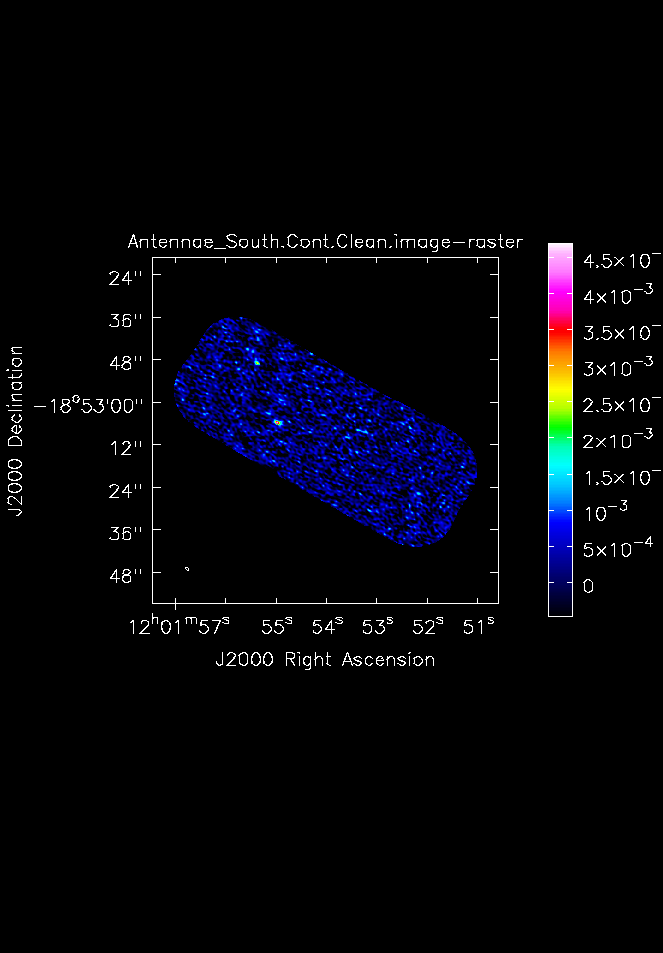
[[File:Antennae_South.Cont.Clean.image.png|200px|thumb|right|Fig. 6. 345 GHz continuum image of the southern mosaic made with imview.]] | [[File:Antennae_South.Cont.Clean.image.png|200px|thumb|right|'''Fig. 6.''' 345 GHz continuum image of the southern mosaic made with {{imview}}.]] | ||
You can determine statistics for the images using the task {{imstat}}: | You can determine statistics for the images using the task {{imstat}}: | ||
| Line 139: | Line 159: | ||
</source> | </source> | ||
From this we find, that for the Northern continuum image the peak is 3. | From this we find, that for the Northern continuum image the peak is 3.80 mJy/beam and the rms is 0.41 mJy/beam. For the Southern continuum we find a peak of 5.23 mJy/beam and an rms of 0.42 mJy/beam. | ||
However, the calculation of the rms comes with a couple of caveats. First, the mosaic primary beam response rolls off toward the edges of the mosaic, as do correspondingly | However, the calculation of the rms comes with a couple of caveats. First, the mosaic primary beam response rolls off toward the edges of the mosaic, as do correspondingly the flux density and rms. Thus if you don't restrict the measurement to areas of full sensitivity, the apparent rms is skewed downward. Second, since there is real emission in the image, the rms will be skewed upward (with the error increasing with brighter emission). Both can be solved by picking boxes that exclude the edges of the mosaic and the real emission. | ||
It is often easier to use either the {{viewer}} directly or | It is often easier to use either the {{viewer}} directly or {{imview}} (a task wrapper for the {{viewer}}; see '''help imview''' for more info) to display the image and then interactively use the region tools to get the statistics. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 150: | Line 170: | ||
</source> | </source> | ||
Adjust colorscale using the middle mouse button (or reassign to different mouse button by clicking on "plus" symbol icon). Next, select the polygon tool by clicking on its symbol with a mouse button. Then draw a region that avoids edges and emission (see example), then double click inside the polygon to have the statistic printed to the screen. We find rms=0. | Adjust colorscale using the middle mouse button (or reassign to a different mouse button by clicking on "plus" symbol icon). Next, select the polygon tool by clicking on its symbol with a mouse button. Then draw a region that avoids edges and emission (see example), then double click inside the polygon to have the statistic printed to the screen. We find rms=0.47 mJy/beam, 15% larger. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 157: | Line 177: | ||
</source> | </source> | ||
We find rms=0. | We find rms=0.50 mJy/beam, 19% larger than what we got from using {{imstat}} with no constraints. | ||
How does this compare to theory? You can find out using the [https://almascience.nrao.edu/call-for-proposals/sensitivity-calculator ALMA sensitivity calculator]. The continuum bandwidth of the Northern mosaic is about 1 GHz and about 0.85 GHz for the Southern mosaic. The number of antennas is about 12 and the time on a single pointing is about 300s. This yields an expected rms of about 1 mJy/beam. However, this needs to be decreased by about a factor of 2.5 near the center of the mosaic due to the hexagonal Nyquist sampling of the mosaic (radial spacing~0.37*HPBW) for a theoretical rms of about 0.4 mJy/beam, in good agreement with observation. | |||
Next make hardcopy plots of the continuum images using | Next make hardcopy plots of the continuum images using {{imview}}. To make the contrast a bit better, set the data range from -1sigma to the peak determined above. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 165: | Line 187: | ||
imview ( | imview ( | ||
raster={'file': 'Antennae_North.Cont.Clean.image', | raster={'file': 'Antennae_North.Cont.Clean.image', | ||
'colorwedge':T,'range':[-0. | 'colorwedge':T,'range':[-0.00048,0.00380]}, | ||
zoom=1, out='Antennae_North.Cont.Clean.image.png') | zoom=1, out='Antennae_North.Cont.Clean.image.png') | ||
imview ( | imview ( | ||
raster={'file': 'Antennae_South.Cont.Clean.image', | raster={'file': 'Antennae_South.Cont.Clean.image', | ||
'colorwedge':T,'range':-0. | 'colorwedge':T,'range':[-0.00050,0.00523]}, | ||
zoom=1, out='Antennae_South.Cont.Clean.image.png') | zoom=1, out='Antennae_South.Cont.Clean.image.png') | ||
</source> | </source> | ||
| Line 175: | Line 197: | ||
==Continuum subtraction == | ==Continuum subtraction == | ||
In these data, the continuum emission is too weak to contaminate the line emission (i.e. the peak continuum emission is less than the rms noise in the spectral line channels). | In these data, the continuum emission is too weak to contaminate the line emission (i.e. the peak continuum emission is less than the rms noise in the spectral line channels). Nevertheless, for illustrative purposes we demonstrate how to subtract the continuum emission in the uv-domain using the task {{uvcontsub}}. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
uvcontsub(vis='Antennae_North.cal.ms',fitspw='0:1~50;120~164',fitorder = 1) | |||
# In CASA | |||
uvcontsub(vis='Antennae_South.cal.ms',fitspw='0:1~30;120~164',fitorder = 1) | |||
</source> | </source> | ||
where: '''fitspw''' gives the line-free channels for each mosaic and '''fitorder'''=1. Higher order fits are not recommended. If you do not have line-free channels on both sides of the line '''fitorder'''=0 is recommended. | where: '''fitspw''' gives the line-free channels for each mosaic and '''fitorder'''=1. Higher order fits are not recommended. If you do not have line-free channels on both sides of the line '''fitorder'''=0 is recommended. The output ms will have .contsub appended to the name. | ||
== CO(3-2) Imaging == | == CO(3-2) Imaging == | ||
[[Image:North_CO3_2_vel.png|thumb|Northern CO(3-2) uv-spectrum in LSRK velocity space.]] | [[Image:North_CO3_2_vel.png|thumb|'''Fig. 7a.''' Northern CO(3-2) uv-spectrum in LSRK velocity space.]] | ||
[[Image:South_CO3_2_vel.png|thumb|Southern CO(3-2) uv-spectrum in LSRK velocity space.]] | [[Image:South_CO3_2_vel.png|thumb|'''Fig. 7b.''' Southern CO(3-2) uv-spectrum in LSRK velocity space.]] | ||
Now we are ready to make cubes of the line emission. The imaging parameters are similar to the continuum except for | Now we are ready to make cubes of the line emission. The imaging parameters are similar to the continuum except for | ||
those dealing with the spectral setup: '''mode, start, width, nchan, restfreq, and outframe''' parameters. When making spectral images you have three choices for the '''mode''' parameter: '''channel''', '''velocity''', and '''frequency'''. Data are taken using constant frequency channels. For spectral line analysis it's often more useful to have constant velocity channels, and this is also the best way to make images of multiple lines with the exact same channelization for later comparison. For '''mode='velocity'''', the desired '''start''' and '''width''' also need to be given in velocity units for the desired output frame. | those dealing with the spectral setup: '''mode, start, width, nchan, restfreq, and outframe''' parameters. When making spectral images you have three choices for the '''mode''' parameter: '''channel''', '''velocity''', and '''frequency'''. Data are taken using constant frequency channels. For spectral line analysis it's often more useful to have constant velocity channels, and this is also the best way to make images of multiple lines with the exact same channelization for later comparison. For '''mode='velocity'''', the desired '''start''' and '''width''' also need to be given in velocity units for the desired output frame. | ||
It is important to note that ALMA does not do on-line Doppler Tracking and the native frame of the data is TOPO. If you do not specify '''outframe''' the output cube will also be in TOPO, which is not very useful for | It is important to note that ALMA does not do on-line Doppler Tracking and the native frame of the data is TOPO. If you do not specify '''outframe''' the output cube will also be in TOPO, which is not very useful for scientific analysis. The Doppler Shift is taken out during the regridding to the desired outframe in {{clean}} or alternatively it can be done separately by the {{cvel}} task which would need to be run before {{clean}}. | ||
To see what velocity parameters you want to set in clean it is useful to make a plot in {{plotms}} in the desired output frame. Note these plots take a little longer because of the frame shift. In order to compare with | To see what velocity parameters you want to set in {{clean}}, it is useful to make a plot in {{plotms}} in the desired output frame. Note these plots take a little longer because of the frame shift. In order to compare with recent SMA data, we chose LSRK, but it should be noted that many papers of this source are in the BARY frame. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 216: | Line 238: | ||
As before, it is very important that you make a clean mask. There are many ways to do this ranging from the complicated to simple. For this example we will make a single clean mask that encompasses the line emission in every channel and apply it to all channels. This is much better than no clean mask, though not quite as good as making an individual mask for each channel. | As before, it is very important that you make a clean mask. There are many ways to do this ranging from the complicated to simple. For this example we will make a single clean mask that encompasses the line emission in every channel and apply it to all channels. This is much better than no clean mask, though not quite as good as making an individual mask for each channel. | ||
<u>Notable parameters | <u>Notable parameters included in the {{clean}} call are:</u> | ||
*'''mode | *'''mode'''='velocity','''outframe'''='LSRK','''restfreq'''='345.79599GHz'. We use 'velocity' mode to set velocity information in the Local Standard of Rest frame (kinematic definition), and use the rest frequency of the CO(3-2) line. | ||
*'''nchan'''=60,start='1300km/s',width='10km/s': To produce a data cube with 60 channels("nchan"=60), starting at 1300km/s and with velocity widths of 10 km/s. That will include all the CO(3-2) emission in both | *'''nchan'''=60,'''start'''='1300km/s','''width'''='10km/s': To produce a data cube with 60 channels ("nchan"=60), starting at 1300km/s and with velocity widths of 10 km/s. That will include all the CO(3-2) emission in both mosaics. | ||
*'''imsize'''= | *'''imsize'''=500,'''cell'''='0.13arcsec'. An image size of 65 arcsec, with pixels of 0.13 arcsec (about one-fifth the minor axis of the synthesized beam). We make the imsize larger than the mosaic to increase the dirty beam image size, as the emission is quite extended along the mosaic. | ||
*''' | *'''weighting'''='briggs','''robust'''=0.5: Weighting to apply to visibilities. We use Briggs' weighting and robustness parameter 0.5 (in between natural and uniform weighting). | ||
*'''niter'''=10000: Maximum number of clean iterations. | *'''niter'''=10000: Maximum number of clean iterations. | ||
*'''threshold=''''5.0mJy': Stop cleaning if the maximum residual is below this value. | *'''threshold=''''5.0mJy': Stop cleaning if the maximum residual is below this value. | ||
The threshold is | The threshold is set to roughly the rms of a single line-free channel for each dataset. | ||
[[Image:North_interact.png|200px|thumb|right|Fig. | [[Image:North_interact.png|200px|thumb|right|'''Fig. 8a.''' Interactive cleaning of northern mosaic. The clean mask is shown in white. In this example we used the same mask for all channels by selecting "all channels" before drawing mask.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 242: | Line 264: | ||
</source> | </source> | ||
Figure 8a shows what the final clean box should look like. Now clean using the green circle arrow. You can watch the progress in the logger. When 100 iterations are done, the viewer will show the residual map for each channel. Cycle through the channels and see whether you are still happy with the clean box in each channel with significant signal. The "erase button" can help you fix mistakes. If necessary adjust. It is often useful to adjust the colorscale with the "plus" symbol icon. To make it go a bit faster, you can increase '''iteration''' interactively, to 200 or 300, but don't overdo it. | |||
These data are pretty severely ''dynamic range'' limited, in part due to the sparse uv coverage. In other words, the noise in bright channels is set by a maximum signal-to-noise. This effectively prevents us from stopping clean based on an rms based '''threshold''' because the effective rms changes as a function of the brightest signal in each channel. Thus, in this case we need to stop clean interactively. Below an approximate number of '''iterations''' is given to help you decide when to quit. NOTE: the '''threshold''' that is set in the CO(3-2) clean commands are about equal to the noise in a line-free channel and is only there to prevent clean running forever if you fail to stop it. You are not intended to clean to this '''threshold'''. | |||
Also, we are going to self-calibrate the data so it's best to be conservative here - it can be difficult in images like these to discern real features from artifacts. If in doubt, don't include it in the clean mask. Keep cleaning and see if the feature becomes weaker. You cannot lose real emission by not masking it, but you can create a brighter feature by masking an artifact. | |||
Stop cleaning after about 900 iterations (Red | Stop cleaning after about 900 iterations (Red X), when the artifacts start to look as bright as the residual. | ||
Inspect the resulting data cube: | Inspect the resulting data cube: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
viewer('Antennae_North. | viewer('Antennae_North.CO3_2Line.Clean.image') | ||
</source> | </source> | ||
==== Southern Mosaic ==== | ==== Southern Mosaic ==== | ||
[[Image: | [[Image:South_interact.png|200px|thumb|right|'''Fig. 8b.''' Interactive cleaning of southern mosaic. The clean mask is shown in white. In this example we used the same mask for all channels by selecting "all channels" before drawing mask.]] | ||
Repeat process for Southern mosaic. | Repeat process for Southern mosaic. | ||
| Line 279: | Line 303: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
viewer('Antennae_South. | viewer('Antennae_South.CO3_2Line.Clean.image') | ||
</source> | </source> | ||
The | For each mosaic use the '''viewer''' polygon tool as described above for the continuum, to find the image statistics in both a line-free channel and the channel with the strongest emission. | ||
The line-free channel rms for the northern and southern mosaics are about 4.0 mJy/beam and 3.5 mJy/beam, respectively. | |||
However in a bright channel this degrades to about 18 mJy/beam and 9 mJy/beam, respectively. This is the aforementioned dynamic range limit. | |||
Using the [https://almascience.nrao.edu/call-for-proposals/sensitivity-calculator ALMA sensitivity calculator], a bandwidth of 10 km/s, dual polarization, 12 antennas, and the time on a single pointing of about 300s yields an rms of about 9.0 mJy/beam. As described above the Nyquist sampled hexagonal mosaic improves this by a factor of about 2.5 for an expected rms noise of about 3.6 mJy. So in line-free channels we are doing close to theoretical. | |||
== Self Calibration == | == Self Calibration == | ||
Next we attempt to self-calibrate the uv-data. The continuum is far | Next we attempt to self-calibrate the uv-data. The continuum is far too weak so we will use the CO(3-2) line emission. The process of self-calibration tries to adjust the data to better match the model image that you give it. That is why it is important to make the model as good as you can by making clean masks. The clean parameter '''calready=T''' (true by default in the commands above) ensured that a uv-model of the resulting image was placed in the model data column. This model can then be used to self-calibrate the data using {{gaincal}}. | ||
Mosaics can be tricky to self-calibrate. The reason is that typically only a few of the pointings may have strong emission. The stronger the signal, the stronger the signal-to-noise will be for a given self-calibration solution interval ('''solint'''). This means that typically only a few fields are strong enough for self-calibration, though it can still bring about overall improvement to apply these solutions to all fields. The fine tuning of each field in the mosaic over small solutions intervals is often impossible. Indeed fine-tuning only a few fields in the mosaic on small time scales can result in a mosaic with dramatically different noise levels as a function of position. Below, we will simply attempt to self-calibrate on the strongest field in the mosaic, obtaining one average solution per original input dataset. | |||
Another wrinkle is the self-calibration of spectral line data. To increase signal-to-noise it is often helpful to limit solutions to the range of strongest line emission. | Another wrinkle is the self-calibration of spectral line data. To increase signal-to-noise it is often helpful to limit solutions to the range of channels containing the strongest line emission. | ||
We will also use '''gaintype='T'''' to tell | We will also use '''gaintype='T'''' to tell CASA to average the polarizations before deriving solutions which gains us a sqrt(2) in sensitivity. | ||
==== Northern Mosaic ==== | ==== Northern Mosaic ==== | ||
[[Image:north_selfchan.png|200px|thumb|right| Plot of the CO(3-2) emission in Field | [[Image:north_selfchan.png|200px|thumb|right|'''Fig. 9a.''' Plot of the CO(3-2) emission in Field 12 of the Northern mosaic.]] | ||
The strongest emission in the Northern mosaic comes from the center pointing, already identified above as '''field=12'''. Below we make a uv-plot of the spectral line emission from this field to chose only the strongest channels to include in the self-calibration. | The strongest emission in the Northern mosaic comes from the center pointing, already identified above as '''field='12''''. Below we make a uv-plot of the spectral line emission from this field to chose only the strongest channels to include in the self-calibration. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='Antennae_North.cal.ms.contsub',spw='',xaxis='channel',yaxis='amp',field='12', | plotms(vis='Antennae_North.cal.ms.contsub',spw='',xaxis='channel',yaxis='amp',field='12', | ||
avgtime='1e8',avgscan=T,coloraxis='field') | avgtime='1e8',avgscan=T,coloraxis='field',plotfile='North_selfchan.png') | ||
</source> | </source> | ||
| Line 313: | Line 340: | ||
# In CASA | # In CASA | ||
plotms(vis='Antennae_North.cal.ms.contsub',spw='',xaxis='time',yaxis='amp',field='12', | plotms(vis='Antennae_North.cal.ms.contsub',spw='',xaxis='time',yaxis='amp',field='12', | ||
avgchannel='167' | avgchannel='167',coloraxis='scan') | ||
</source> | </source> | ||
Zooming in on each dataset we see that they are about 1 hour long (you can also check the | Zooming in on each dataset we see that they are about 1 hour long (you can also check the {{listobs}} output), and that each dataset has 2 or 3 observations of field 12. Each time, it's observed for about 25 seconds. We set '''solint='3600s'''' to get one solution per dataset. | ||
[[Image:north_pcal1_phase.png|200px|thumb|right| Phase-only self-calibration solutions for the Northern mosaic.]] | [[Image:north_pcal1_phase.png|200px|thumb|right|'''Fig. 9b.''' Phase-only self-calibration solutions for the Northern mosaic.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 338: | Line 365: | ||
</source> | </source> | ||
Apply the solutions to all fields and channels | Apply the solutions to all fields and channels with {{applycal}}, which will overwrite the corrected data column: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 346: | Line 373: | ||
</source> | </source> | ||
Re-image the data with the selfcal applied. | Re-image the data with the selfcal applied. Remember that {{clean}} always uses the corrected data column if it exists: | ||
[[Image:North_interact2.png|200px|thumb|right|'''Fig. 9c.''' Interactive cleaning of Northern mosaic after self-cal. The clean mask is shown in white. In this example we used the same mask for all channels by selecting "all channels" before drawing mask.]] | |||
<source lang="python"> | <source lang="python"> | ||
| Line 357: | Line 386: | ||
nchan=70,start='1200km/s',width='10km/s', | nchan=70,start='1200km/s',width='10km/s', | ||
imagermode='mosaic', | imagermode='mosaic', | ||
imsize= | imsize=500,cell='0.13arcsec',minpb=0.2, | ||
interactive=T,mask='Antennae_North.CO3_2Line.Clean.mask', | interactive=T,mask='Antennae_North.CO3_2Line.Clean.mask', | ||
weighting='briggs',robust=0.5, | weighting='briggs',robust=0.5, | ||
| Line 365: | Line 394: | ||
After 600 iterations or so, inspect each channel to see if there is more emission that needs to be included in the mask. Remember to select the "all channels" toggle. Stop after 1200 or so iterations. | After 600 iterations or so, inspect each channel to see if there is more emission that needs to be included in the mask. Remember to select the "all channels" toggle. Stop after 1200 or so iterations. | ||
Compare the un-selfcal'ed and | Compare the un-selfcal'ed and selfcal'ed data to determine if there has been improvement. | ||
<source lang="python"> | |||
# In CASA | |||
imview (raster=[{'file': 'Antennae_North.CO3_2Line.Clean.image', | |||
'range': [-0.07,0.4]}, | |||
{'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image', | |||
'range': [-0.07,0.4]}], | |||
zoom={'channel': 43}) | |||
</source> | |||
Then in the viewer, select the blink toggle in the "tapedeck", now the tapedeck buttons cycle between the two images at the selected channel. To change the channel toggle back to "normal", change the channel number, and then toggle back to "blink". | |||
To make the figure shown in 9.d, you can select the "p wrench" icon and change the number of panels in x to 2. You will need to drag your viewer window rather wide to get both panels to fit well. If you want to compare other channels, you need to toggle blink off, then use the tapedeck to move to the new channel of interest, and then turn blink back on to see both cubes again. | |||
Note: while in "blink" mode, statistics will be for the final image in the stack. In "normal" mode, all open raster images will have their statistics reported. | |||
What you should notice is that the peak flux density has increased substantially while the rms noise is about the same. This is reasonable for this self-calibration case because we have mostly adjusted relative position offsets between the datasets and not taken out any short-term phase variations which would reduce the rms. | |||
Note that attempts to push the selfcal to shorter solint (for example to each scan of field=12) did not yield additional improvement. | |||
[[Image:north_ch43compare.png|600px|thumb|center| '''Fig 9.d.''' Comparison of channel 43 before selfcal (left) and after selfcal (right).]] | |||
==== Southern Mosaic ==== | ==== Southern Mosaic ==== | ||
[[Image:south_selfchan.png|200px|thumb|right|'''Fig. 10a.''' Plot of the CO(3-2) emission in Field 7 of the Southern mosaic.]] | |||
Now repeat the self-calibration process for the Southern mosaic. It is a bit tougher in this case to pick the best field. The most compact bright emission is toward field id 11 in the [[AntennaeBand7#Science_Target_Overview | Overview]]. Adjusting for the renumbering of field ids after the final calibration split, this becomes field='7'. | |||
From the following plots we can assess the brightest channels to pick and see that the timing in the 6 Southern datasets is similar to the North, with each lasting about 1 hour. | |||
<source lang="python"> | <source lang="python"> | ||
| Line 380: | Line 435: | ||
avgchannel='167',avgscan=T,coloraxis='scan') | avgchannel='167',avgscan=T,coloraxis='scan') | ||
</source> | </source> | ||
[[Image:south_pcal1_phase.png|200px|thumb|right|'''Fig. 10b.''' Phase-only self-calibration solutions for the Southern mosaic.]] | |||
<source lang="python"> | <source lang="python"> | ||
| Line 402: | Line 459: | ||
gaintable=['south_self_1.pcal'],calwt=F) | gaintable=['south_self_1.pcal'],calwt=F) | ||
</source> | </source> | ||
[[Image:South_interact2.png|200px|thumb|right|'''Fig. 10c.''' Interactive cleaning of Southern mosaic after self-cal. The clean mask is shown in white. In this example we used the same mask for all channels by selecting "all channels" before drawing mask.]] | |||
<source lang="python"> | <source lang="python"> | ||
| Line 418: | Line 477: | ||
</source> | </source> | ||
After 600 iterations or so, inspect each channel to see if there is more emission that needs to be included in the mask. Remember to select the "all channels" toggle. Stop after 2200 or so iterations. | |||
As for the Northern mosaic, you can compare the before and after self-cal images: | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
imview (raster=[{'file': 'Antennae_South.CO3_2Line.Clean.image', | |||
'range': [-0.05,0.2]}, | |||
{'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image', | |||
'range': [-0.05,0.2]}], | |||
zoom={'channel': 31}) | |||
</source> | </source> | ||
[[Image:South_ch31compare.png|600px|thumb|center| '''Fig. 10d.''' Comparison of channel 31 before selfcal (left) and after selfcal (right).]] | |||
== Image Analysis == | == Image Analysis == | ||
| Line 462: | Line 496: | ||
==== Moment Maps ==== | ==== Moment Maps ==== | ||
[[Image:Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.png|200px|thumb|right|'''Fig. 11a.''' The CO(3-2) integrated intensity map (moment 0) of the Northern mosaic.]] | |||
[[Image:Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png|200px|thumb|right|'''Fig. 11b.''' The CO(3-2) velocity field (moment 1) of the Northern mosaic.]] | |||
[[Image:Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png|200px|thumb|right|'''Fig. 11c.''' The CO(3-2) velocity dispersion (moment 2) of the Northern mosaic.]] | |||
[[Image:Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.png|200px|thumb|right|'''Fig. 12a.''' The CO(3-2) integrated intensity map (moment 0) of the Southern mosaic.]] | |||
[[Image:Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png|200px|thumb|right|'''Fig. 12b.''' The CO(3-2) velocity field (moment 1) of the Southern mosaic.]] | |||
[[Image:Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png|200px|thumb|right|'''Fig. 12c.''' The CO(3-2) velocity dispersion (moment 2) of the Southern mosaic.]] | |||
Next we will make moment maps for the CO(3-2) emission: Moment 0 is the integrated intensity; Moment 1 is the intensity weighted velocity field; and Moment 2 is the intensity weighted velocity dispersion. | |||
Above we determined the rms noise levels for both the North and South mosaics in both a line-free and a line-bright channel. We want to limit the channel range of the moment calculations to those channels with significant emission. One good way to do this is to open the cube in the viewer overlaid with 3sigma contours, with sigma corresponding to the line-free rms. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
imview(raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image', | |||
'range': [-0.04,0.4]}, | |||
contour={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image', | |||
'levels': [0.004],'unit': 5}) | |||
</source> | </source> | ||
We find a channel range for significant emission of 33~63. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
imview(raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image', | |||
'range': [-0.04,0.4]}, | |||
contour={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image', | |||
'levels': [0.0035],'unit': 5}) | |||
</source> | |||
We find a channel range for significant emission of 12~64. | |||
immoments(' | For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity. | ||
<source lang="python"> | |||
# In CASA | |||
immoments('Antennae_North.CO3_2Line.Clean.pcal1.image', | |||
moments=[0],chans='33~63', | |||
outfile='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0') | |||
</source> | </source> | ||
<source lang="python"> | |||
# In CASA | |||
immoments('Antennae_South.CO3_2Line.Clean.pcal1.image', | |||
moments=[0], chans='12~64', | |||
outfile='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0') | |||
</source> | |||
For higher order moments it is very important to set a conservative flux threshold. Typically something like 3sigma, using sigma from a bright line channel works well. We do this with the '''mask''' parameter in the commands below. When making multiple moments, {{immoments}} uses the '''out''' parameter as a base and appends an appropriate ending. | |||
<source lang="python"> | |||
# In CASA | |||
immoments('Antennae_North.CO3_2Line.Clean.pcal1.image', | |||
moments=[1,2], chans='33~63', | |||
mask='Antennae_North.CO3_2Line.Clean.pcal1.image>0.018*3', | |||
outfile='Antennae_North.CO3_2Line.Clean.pcal1.image.mom') | |||
</source> | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
immoments('Antennae_South.CO3_2Line.Clean.pcal1.image', | |||
moments=[1,2], chans='12~64', | |||
mask=' Antennae_South.CO3_2Line.Clean.pcal1.image>0.009*3', | |||
outfile='Antennae_South.CO3_2Line.Clean.pcal1.image.mom') | |||
</source> | |||
Next we can create the six moment map figures (11abc, 12abc) from these images with '''imview'''. | |||
imview ( | <source lang="python"> | ||
# In CASA | |||
os.system('rm -f Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.png') | |||
imview (raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image.mom0', | |||
'colorwedge':T,'scaling': -0.5}, | |||
zoom=1,out='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.png') | |||
os.system('rm -f Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png') | |||
imview (raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord', | |||
'colorwedge':T}, | |||
zoom=1,out='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png') | |||
imview ( | os.system('rm -f Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png') | ||
imview (raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord', | |||
'colorwedge':T}, | |||
zoom=1,out='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png') | |||
== | os.system('rm -f Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.png') | ||
imview (raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image.mom0', | |||
'colorwedge':T,'scaling': -0.5}, | |||
zoom=1,out='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.png') | |||
os.system('rm -f Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png') | |||
imview (raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord', | |||
'colorwedge':T}, | |||
zoom=1,out='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png') | |||
os.system('rm -f Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png') | |||
imview (raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord', | |||
'colorwedge':T}, | |||
zoom=1,out='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png') | |||
</source> | |||
==Exporting to Fits == | ==== Exporting Images to Fits ==== | ||
If you want to analyze the data using another software package it is easy to convert from CASA format to FITS. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
exportfits(imagename='Antennae_North. | exportfits(imagename='Antennae_North.Cont.Clean.image', | ||
fitsimage='Antennae_North. | fitsimage='Antennae_North.Cont.Clean.image.fits') | ||
exportfits(imagename='Antennae_North. | exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image', | ||
fitsimage='Antennae_North. | fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.fits') | ||
exportfits(imagename='Antennae_North. | exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0', | ||
fitsimage='Antennae_North. | fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.fits') | ||
exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord', | |||
fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.fits') | |||
exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord', | |||
fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.fits') | |||
</source> | </source> | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
exportfits(imagename='Antennae_South. | exportfits(imagename='Antennae_South.Cont.Clean.image', | ||
fitsimage='Antennae_South. | fitsimage='Antennae_South.Cont.Clean.image.fits') | ||
exportfits(imagename='Antennae_South. | exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image', | ||
fitsimage='Antennae_South. | fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.fits') | ||
exportfits(imagename='Antennae_South. | exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0', | ||
fitsimage='Antennae_South. | fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.fits') | ||
exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord', | |||
fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.fits') | |||
exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord', | |||
fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.fits') | |||
</source> | </source> | ||
Although "fits format" is supposed to be a standard, in fact most packages expect slightly different things from a fits image. If you are having difficulty, try setting velocity=T and/or dropstokes=T. | |||
==Comparison with previous SMA CO(3-2) data== | |||
We compare with SMA CO(3-2) data (Ueda, Iono, Petitpas et al., submitted to ApJ). Figure 13 shows a comparison plot between the moment 0 maps of ALMA and SMA data using the {{viewer}}. The fluxes, peak locations, and large scale structure are consistent. Both southern and northern components have been combined. | |||
[[Image: alma_sma_compare.png|500px|thumb|center| '''Fig. 13.''' The CO(3-2) total intensity map (moment 0) comparison with SMA data. Colour image is ALMA data, combining southern and northern mosaics. Contours show SMA data (Ueda, Iono, Petitpas et al., submitted to ApJ).]] | |||
{{Checked 3.3.0}} | |||
Latest revision as of 20:05, 16 May 2012
Overview
This section of the AntennaeBand7 CASA Guide covers the imaging of the continuum and spectral line data. It begins where the Antennae Band7 - Calibration section left off. If you completed the Calibration section of the guide, then you already have the calibrated data sets. If you did not complete the Calibration section, then you can download the calibrated uv-data from the region closest to your location:
North America Europe East Asia
Then download the file 'Antennae_Band7_CalibratedData.tgz' to obtain the calibrated uvdata. Once the download has finished, unpack the file using
> 'tar -xvzf Antennae_Band7_CalibratedData.tgz'
once it's unpacked, change directory (cd) to Antennae_Band7_CalibratedData. You should have Antennae_South.cal.ms and Antennae_North.cal.ms in your working directory. Then start CASA with the 'casapy' command. Be sure you are using version casapy-stable r19407 or later.
Imaging Mosaics
If you are unfamiliar with the basic concepts of deconvolution and clean, pause here and review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/wilner_synthesis10.pdf
Mosaics like other kinds of images are created in the CASA task clean. To invoke mosaic mode, you simply set the parameter imagermode='mosaic'. The default subparameter ftmachine='mosaic' is then automatically set. This is a joint deconvolution algorithm that works in the uv-plane. A convolution of the primary beam patterns for each pointing in the mosaic is created: the primary beam response function. The corresponding image of the mosaic response function will be called <imagename>.flux.pbcoverage and <imagename>.flux (where the latter differs from the former only if the sensitivity of each field in the mosaic varies).
Well-sampled mosaics like the patterns observed for the Northern and Southern Antennae mosaics shown on the Overview page are well-suited to joint deconvolution. However, if you have a poorly-sampled, or very irregular mosaic you may need to use the slower ftmachine='ft' which combines data in the image plane, similar to what is done in other packages like miriad.
Additionally, for mosaics it is essential to pick the center of the region to be imaged explicitly using the phasecenter parameter. Otherwise it will default to the first pointing included in the field parameter -- since this is often at one corner of the mosaic, the image will not be centered. For the Northern mosaic, the center pointing corresponds to field id=12. Note that during the final split in the calibration section that selected only the Antennae fields, the field ids were renumbered, so that the original centers (shown in the Overview) have changed: field id 14 becomes 12 for the Northern mosaic and field 18 becomes 15 for the Southern mosaic. You can also set an explicit coordinate (see the clean help for syntax).
If you want to learn more about mosaicing, pause here and review for example http://www.aoc.nrao.edu/events/synthesis/2010/lectures/jott-mosaicking-school-04.pdf
Continuum Imaging
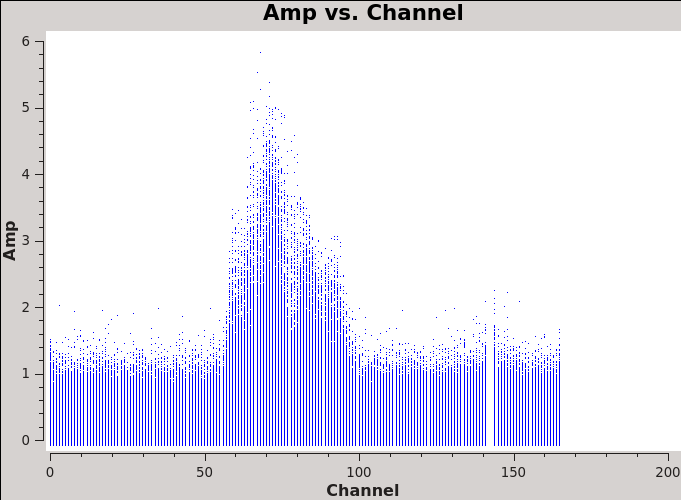
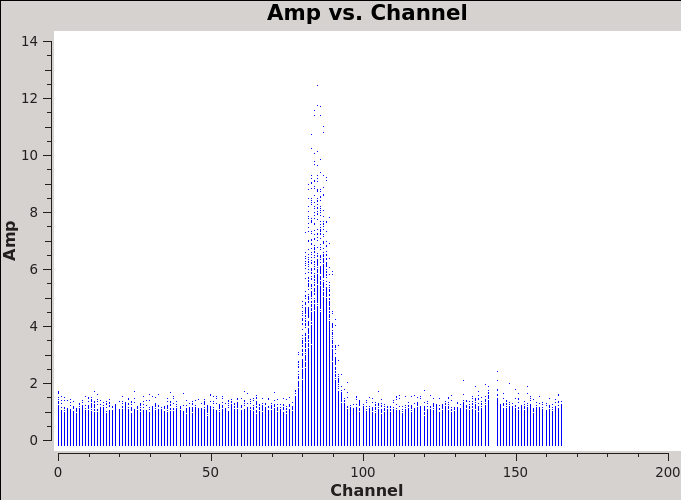
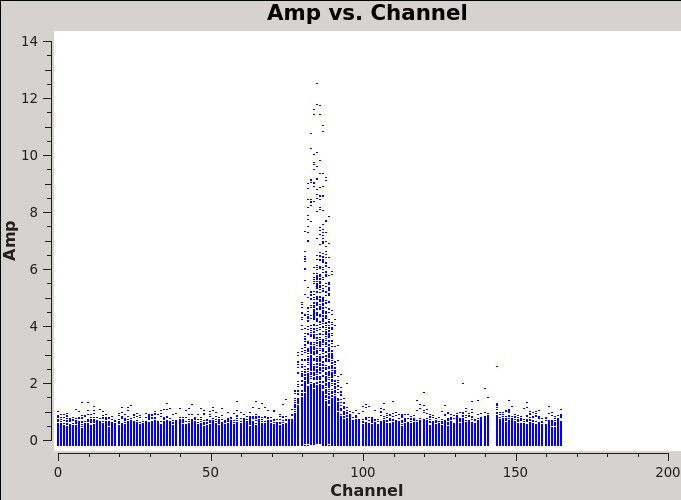
We will make 345 GHz continuum images for the two regions covered by the mosaics. We use the task clean over the channels that are free of the line emission; we avoid the edge channels which tend to be noisier due to bandpass rolloff effects. The line-free channels are found by plotting the average spectrum (all fields). We find the CO(3-2) line from channels 50 to 100 in the southern mosaic (Figure 1), and from 70 to 100 in the northern mosaic (Figure 2).
# In CASA
plotms(vis='Antennae_North.cal.ms',xaxis='channel',yaxis='amp',
avgtime='1e8',avgscan=T,plotfile='Antennae_North-AMPvsCH.png')
# In CASA
plotms(vis='Antennae_South.cal.ms',xaxis='channel',yaxis='amp',
avgtime='1e8',avgscan=T,plotfile='Antennae_South-AMPvsCH.png')
The avgtime is set to a large value so that it averages over all the integrations, and avgscan is set to allow averaging of the different scans.
Next we create continuum images from the line-free channels.
Northern Continuum Mosaic
For illustrative purposes we first make a dirty image to see if there is emission and what the exact beam size is. It should be on the order of 1" but this will vary a bit according to the uv-coverage in the actual data. We will start with a cell size of 0.2" to oversample the beam by a factor of 5. The imsize needs to be large enough given the cell size to comfortably encompass the mosaic. From the mosaic footprints shown in the overview, we can see that the Northern mosaic imsize needs to be about 1 arcmin. With 0.2" pixels, this requires a imsize=300.
Other essential clean parameters for this case include:
- vis='Antennae_North.cal.ms': The calibrated dataset on the science target. clean will always use the CORRECTED DATA column (if it exists).
- imagename='Antennae_North.Cont.Dirty': The base name of the output images:
- <imagename>.image # the final restored image
- <imagename>.flux # the effective primary beam response (e.g. for pbcor)
- <imagename>.flux.pbcoverage # the primary beam coverage (ftmachine=’mosaic’ only)
- <imagename>.model # the model image
- <imagename>.residual # the residual image
- <imagename>.psf # the synthesized (dirty) beam
- spw='0:1~50;120~164': To specify only the line-free channels of spectral window 0.
- mode='mfs': Multi-Frequency Synthesis: The default mode, which produces one image from all the specified data combined. This will grid each channel independently before imaging. For wide bandwidths this will give a far superior result compared to averaging the channels and then imaging.
- restfreq='345.79599GHz': The rest frequency of CO(3-2) can be found with splatalogue.
- niter=0: Maximum number of clean iterations -- niter=0 will do no cleaning
# In CASA
os.system('rm -rf Antennae_North.Cont.Dirty*')
clean(vis='Antennae_North.cal.ms',imagename='Antennae_North.Cont.Dirty',
field='',phasecenter='12',
mode='mfs',restfreq='345.79599GHz',
spw='0:1~50;120~164',
imagermode='mosaic',
imsize=300,cell='0.2arcsec',
interactive=F,niter=0)
The reported beam size is about 1.27" x 0.71" P.A. 86.142 degrees.
# In CASA
viewer('Antennae_North.Cont.Dirty.image')
If you are unfamiliar with the CASA viewer it is a good idea to pause here and watch the demo at http://casa.nrao.edu/CasaViewerDemo/casaViewerDemo.html
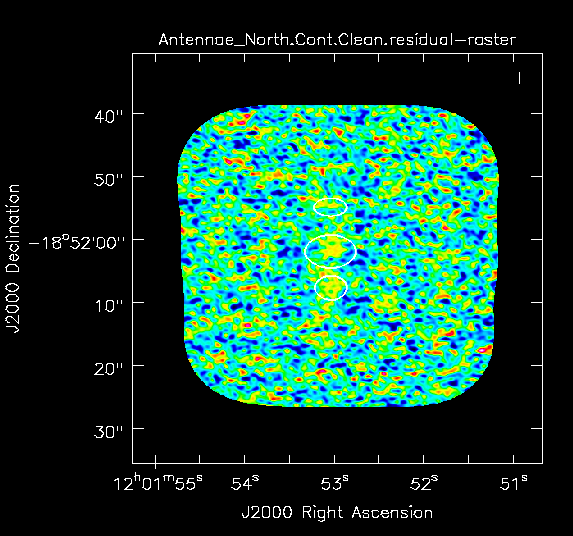
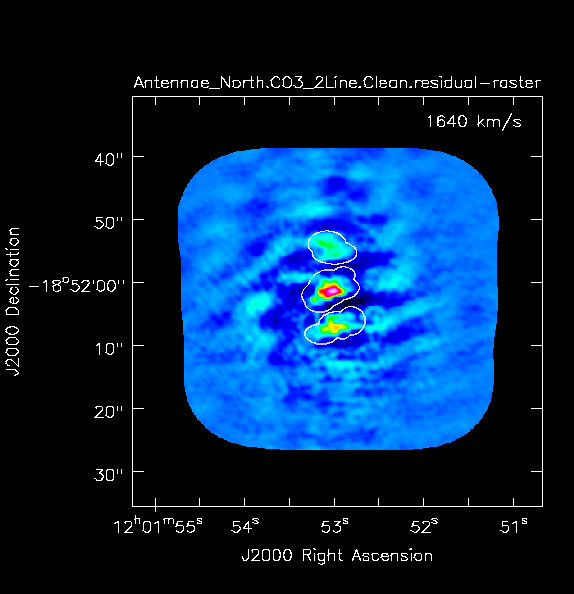
Yes there is definitely a detection in the vicinity of the Northern nucleus (see Figs. 4 and 5 below). Using the square region icon, and drawing a box near but not including the emission, we find the rms noise is about 0.4 mJy/beam in the dirty image.
Next we switch to refined values of cell=0.13" and imsize=500 based on the observed beam size. We also switch to interactive mode so that you can create a clean mask using the polygon tool (note you need to double click inside the polygon region to activate the mask). See TWHya casaguide for a more complete description of interactive clean and mask creation.
- niter=1000: Maximum number of clean iterations -- we will stop interactively
- threshold='0.4mJy': Stop cleaning if the maximum residual is below this value (the dirty rms noise)
- interactive=T: Clean will be periodically interrupted to show the residual clean image. Interactive clean mask can be made. If no mask is created, no cleaning is done.
# In CASA
os.system('rm -rf Antennae_North.Cont.Clean*')
clean(vis='Antennae_North.cal.ms',imagename='Antennae_North.Cont.Clean',
field='',phasecenter='12',
mode='mfs',restfreq='345.79599GHz',
spw='0:1~50;120~164',
imagermode='mosaic',
imsize=500,cell='0.13arcsec',
interactive=T,
niter=1000, threshold='0.4mJy')
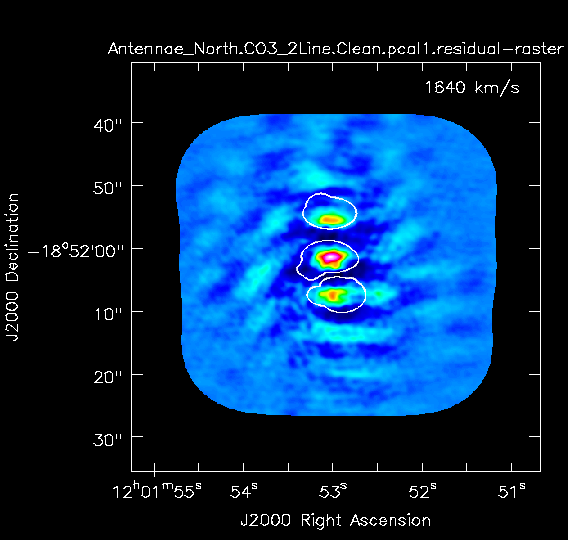
The residuals are "noise-like" after only 100 iterations (see Fig. 3a), so hit the red X symbol in the interactive window to stop cleaning here.
Note that if you run the clean task again with the same imagename, without deleting the existing <imagename>.* files, clean assumes that you want to continue cleaning the existing images. We put an rm command before the clean command to guard against this, but sometimes this is what you want.
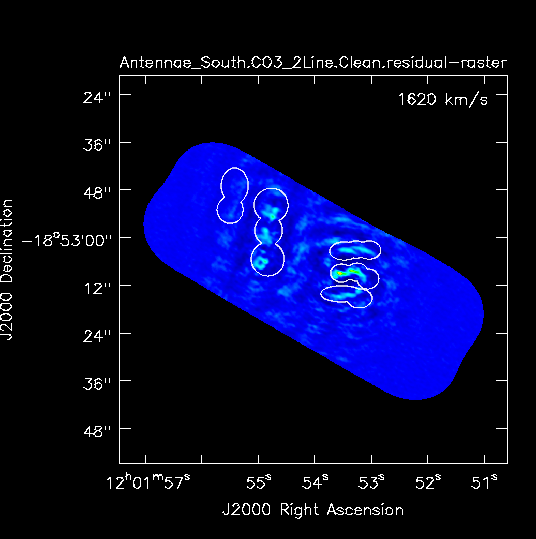
Southern Continuum Mosaic
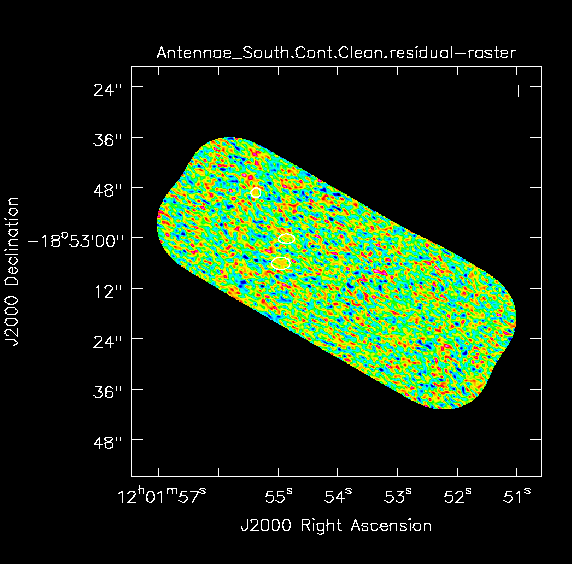
For the southern mosaic we modify the phasecenter, mosaic imsize, and the line-free channels (spw) to be consistent for this mosaic. We also bypass the dirty image step we did above for the Northern mosaic. As above, the mosaic size can be judged from here. We expect the beam to be about the same and use the same cell size. During clean, we repeat the process of making a clean mask.
# In CASA
os.system('rm -rf Antennae_South.Cont.Clean*')
clean(vis='Antennae_South.cal.ms',imagename='Antennae_South.Cont.Clean',
field='',phasecenter='15',
mode='mfs',restfreq='345.79599GHz',
spw='0:1~30;120~164',
imagermode='mosaic',
imsize=750,cell='0.13arcsec',
interactive=T,
niter=1000, threshold='0.4mJy')
The beam size reported in the logger for the Southern mosaic: 1.11"x 0.71", and P.A.=65 deg; which is a little better than the beam for the North owing to better uv-coverage.
Stop after 100 iterations (Fig. 3b).
Image Statistics
You can determine statistics for the images using the task imstat:
# In CASA
imstat('Antennae_North.Cont.Clean.image')
imstat('Antennae_South.Cont.Clean.image')
From this we find, that for the Northern continuum image the peak is 3.80 mJy/beam and the rms is 0.41 mJy/beam. For the Southern continuum we find a peak of 5.23 mJy/beam and an rms of 0.42 mJy/beam.
However, the calculation of the rms comes with a couple of caveats. First, the mosaic primary beam response rolls off toward the edges of the mosaic, as do correspondingly the flux density and rms. Thus if you don't restrict the measurement to areas of full sensitivity, the apparent rms is skewed downward. Second, since there is real emission in the image, the rms will be skewed upward (with the error increasing with brighter emission). Both can be solved by picking boxes that exclude the edges of the mosaic and the real emission.
It is often easier to use either the viewer directly or imview (a task wrapper for the viewer; see help imview for more info) to display the image and then interactively use the region tools to get the statistics.
# In CASA
viewer('Antennae_North.Cont.Clean.image')
Adjust colorscale using the middle mouse button (or reassign to a different mouse button by clicking on "plus" symbol icon). Next, select the polygon tool by clicking on its symbol with a mouse button. Then draw a region that avoids edges and emission (see example), then double click inside the polygon to have the statistic printed to the screen. We find rms=0.47 mJy/beam, 15% larger.
# In CASA
viewer('Antennae_South.Cont.Clean.image')
We find rms=0.50 mJy/beam, 19% larger than what we got from using imstat with no constraints.
How does this compare to theory? You can find out using the ALMA sensitivity calculator. The continuum bandwidth of the Northern mosaic is about 1 GHz and about 0.85 GHz for the Southern mosaic. The number of antennas is about 12 and the time on a single pointing is about 300s. This yields an expected rms of about 1 mJy/beam. However, this needs to be decreased by about a factor of 2.5 near the center of the mosaic due to the hexagonal Nyquist sampling of the mosaic (radial spacing~0.37*HPBW) for a theoretical rms of about 0.4 mJy/beam, in good agreement with observation.
Next make hardcopy plots of the continuum images using imview. To make the contrast a bit better, set the data range from -1sigma to the peak determined above.
# In CASA
imview (
raster={'file': 'Antennae_North.Cont.Clean.image',
'colorwedge':T,'range':[-0.00048,0.00380]},
zoom=1, out='Antennae_North.Cont.Clean.image.png')
imview (
raster={'file': 'Antennae_South.Cont.Clean.image',
'colorwedge':T,'range':[-0.00050,0.00523]},
zoom=1, out='Antennae_South.Cont.Clean.image.png')
Continuum subtraction
In these data, the continuum emission is too weak to contaminate the line emission (i.e. the peak continuum emission is less than the rms noise in the spectral line channels). Nevertheless, for illustrative purposes we demonstrate how to subtract the continuum emission in the uv-domain using the task uvcontsub.
# In CASA
uvcontsub(vis='Antennae_North.cal.ms',fitspw='0:1~50;120~164',fitorder = 1)
# In CASA
uvcontsub(vis='Antennae_South.cal.ms',fitspw='0:1~30;120~164',fitorder = 1)
where: fitspw gives the line-free channels for each mosaic and fitorder=1. Higher order fits are not recommended. If you do not have line-free channels on both sides of the line fitorder=0 is recommended. The output ms will have .contsub appended to the name.
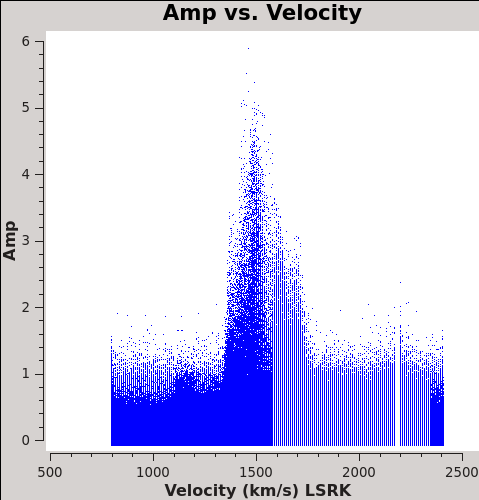
CO(3-2) Imaging
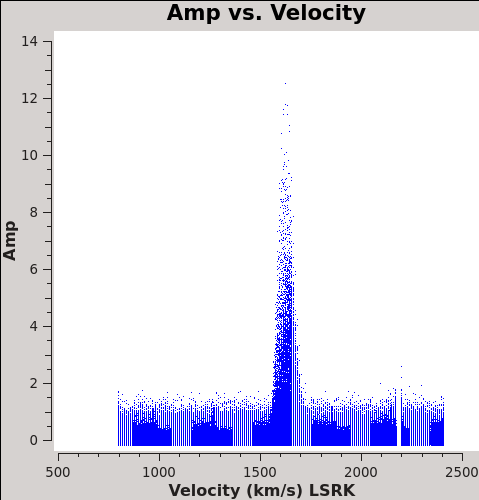
Now we are ready to make cubes of the line emission. The imaging parameters are similar to the continuum except for those dealing with the spectral setup: mode, start, width, nchan, restfreq, and outframe parameters. When making spectral images you have three choices for the mode parameter: channel, velocity, and frequency. Data are taken using constant frequency channels. For spectral line analysis it's often more useful to have constant velocity channels, and this is also the best way to make images of multiple lines with the exact same channelization for later comparison. For mode='velocity', the desired start and width also need to be given in velocity units for the desired output frame.
It is important to note that ALMA does not do on-line Doppler Tracking and the native frame of the data is TOPO. If you do not specify outframe the output cube will also be in TOPO, which is not very useful for scientific analysis. The Doppler Shift is taken out during the regridding to the desired outframe in clean or alternatively it can be done separately by the cvel task which would need to be run before clean.
To see what velocity parameters you want to set in clean, it is useful to make a plot in plotms in the desired output frame. Note these plots take a little longer because of the frame shift. In order to compare with recent SMA data, we chose LSRK, but it should be noted that many papers of this source are in the BARY frame.
# In CASA
plotms(vis='Antennae_North.cal.ms.contsub/',xaxis='velocity',yaxis='amp',
avgtime='1e8',avgscan=T,transform=T,freqframe='LSRK',
restfreq='345.79599GHz',plotfile='North_CO3_2_vel.png')
# In CASA
plotms(vis='Antennae_South.cal.ms.contsub',xaxis='velocity',yaxis='amp',
avgtime='1e8',avgscan=T,transform=T,freqframe='LSRK',
restfreq='345.79599GHz',plotfile='South_CO3_2_vel.png')
Northern Mosaic
As before, it is very important that you make a clean mask. There are many ways to do this ranging from the complicated to simple. For this example we will make a single clean mask that encompasses the line emission in every channel and apply it to all channels. This is much better than no clean mask, though not quite as good as making an individual mask for each channel.
Notable parameters included in the clean call are:
- mode='velocity',outframe='LSRK',restfreq='345.79599GHz'. We use 'velocity' mode to set velocity information in the Local Standard of Rest frame (kinematic definition), and use the rest frequency of the CO(3-2) line.
- nchan=60,start='1300km/s',width='10km/s': To produce a data cube with 60 channels ("nchan"=60), starting at 1300km/s and with velocity widths of 10 km/s. That will include all the CO(3-2) emission in both mosaics.
- imsize=500,cell='0.13arcsec'. An image size of 65 arcsec, with pixels of 0.13 arcsec (about one-fifth the minor axis of the synthesized beam). We make the imsize larger than the mosaic to increase the dirty beam image size, as the emission is quite extended along the mosaic.
- weighting='briggs',robust=0.5: Weighting to apply to visibilities. We use Briggs' weighting and robustness parameter 0.5 (in between natural and uniform weighting).
- niter=10000: Maximum number of clean iterations.
- threshold='5.0mJy': Stop cleaning if the maximum residual is below this value.
The threshold is set to roughly the rms of a single line-free channel for each dataset.
# In CASA
os.system('rm -rf Antennae_North.CO3_2Line*')
clean(vis='Antennae_North.cal.ms.contsub',
imagename='Antennae_North.CO3_2Line.Clean',
spw='0',field='',phasecenter='12',
mode='velocity',outframe='LSRK',restfreq='345.79599GHz',
nchan=70,start='1200km/s',width='10km/s',
imagermode='mosaic',
imsize=500,cell='0.13arcsec',minpb=0.2,
interactive=T,
weighting='briggs',robust=0.5,
niter=10000, threshold='5.0mJy')
Figure 8a shows what the final clean box should look like. Now clean using the green circle arrow. You can watch the progress in the logger. When 100 iterations are done, the viewer will show the residual map for each channel. Cycle through the channels and see whether you are still happy with the clean box in each channel with significant signal. The "erase button" can help you fix mistakes. If necessary adjust. It is often useful to adjust the colorscale with the "plus" symbol icon. To make it go a bit faster, you can increase iteration interactively, to 200 or 300, but don't overdo it.
These data are pretty severely dynamic range limited, in part due to the sparse uv coverage. In other words, the noise in bright channels is set by a maximum signal-to-noise. This effectively prevents us from stopping clean based on an rms based threshold because the effective rms changes as a function of the brightest signal in each channel. Thus, in this case we need to stop clean interactively. Below an approximate number of iterations is given to help you decide when to quit. NOTE: the threshold that is set in the CO(3-2) clean commands are about equal to the noise in a line-free channel and is only there to prevent clean running forever if you fail to stop it. You are not intended to clean to this threshold.
Also, we are going to self-calibrate the data so it's best to be conservative here - it can be difficult in images like these to discern real features from artifacts. If in doubt, don't include it in the clean mask. Keep cleaning and see if the feature becomes weaker. You cannot lose real emission by not masking it, but you can create a brighter feature by masking an artifact.
Stop cleaning after about 900 iterations (Red X), when the artifacts start to look as bright as the residual.
Inspect the resulting data cube:
# In CASA
viewer('Antennae_North.CO3_2Line.Clean.image')
Southern Mosaic
Repeat process for Southern mosaic.
# In CASA
os.system('rm -rf Antennae_South.CO3_2Line*')
clean(vis='Antennae_South.cal.ms.contsub',
imagename='Antennae_South.CO3_2Line.Clean',
spw='0',field='',phasecenter='15',
mode='velocity',outframe='LSRK',restfreq='345.79599GHz',
nchan=70,start='1200km/s',width='10km/s',
imagermode='mosaic',
imsize=750,cell='0.13arcsec',minpb=0.2,
interactive=T,
weighting='briggs',robust=0.5,
niter=10000, threshold='5.0mJy')
Clean about 1200 iterations before stopping.
Inspect the resulting data cube:
# In CASA
viewer('Antennae_South.CO3_2Line.Clean.image')
For each mosaic use the viewer polygon tool as described above for the continuum, to find the image statistics in both a line-free channel and the channel with the strongest emission.
The line-free channel rms for the northern and southern mosaics are about 4.0 mJy/beam and 3.5 mJy/beam, respectively. However in a bright channel this degrades to about 18 mJy/beam and 9 mJy/beam, respectively. This is the aforementioned dynamic range limit.
Using the ALMA sensitivity calculator, a bandwidth of 10 km/s, dual polarization, 12 antennas, and the time on a single pointing of about 300s yields an rms of about 9.0 mJy/beam. As described above the Nyquist sampled hexagonal mosaic improves this by a factor of about 2.5 for an expected rms noise of about 3.6 mJy. So in line-free channels we are doing close to theoretical.
Self Calibration
Next we attempt to self-calibrate the uv-data. The continuum is far too weak so we will use the CO(3-2) line emission. The process of self-calibration tries to adjust the data to better match the model image that you give it. That is why it is important to make the model as good as you can by making clean masks. The clean parameter calready=T (true by default in the commands above) ensured that a uv-model of the resulting image was placed in the model data column. This model can then be used to self-calibrate the data using gaincal.
Mosaics can be tricky to self-calibrate. The reason is that typically only a few of the pointings may have strong emission. The stronger the signal, the stronger the signal-to-noise will be for a given self-calibration solution interval (solint). This means that typically only a few fields are strong enough for self-calibration, though it can still bring about overall improvement to apply these solutions to all fields. The fine tuning of each field in the mosaic over small solutions intervals is often impossible. Indeed fine-tuning only a few fields in the mosaic on small time scales can result in a mosaic with dramatically different noise levels as a function of position. Below, we will simply attempt to self-calibrate on the strongest field in the mosaic, obtaining one average solution per original input dataset.
Another wrinkle is the self-calibration of spectral line data. To increase signal-to-noise it is often helpful to limit solutions to the range of channels containing the strongest line emission.
We will also use gaintype='T' to tell CASA to average the polarizations before deriving solutions which gains us a sqrt(2) in sensitivity.
Northern Mosaic
The strongest emission in the Northern mosaic comes from the center pointing, already identified above as field='12'. Below we make a uv-plot of the spectral line emission from this field to chose only the strongest channels to include in the self-calibration.
# In CASA
plotms(vis='Antennae_North.cal.ms.contsub',spw='',xaxis='channel',yaxis='amp',field='12',
avgtime='1e8',avgscan=T,coloraxis='field',plotfile='North_selfchan.png')
Next remind ourselves of the timing of the various observations.
# In CASA
plotms(vis='Antennae_North.cal.ms.contsub',spw='',xaxis='time',yaxis='amp',field='12',
avgchannel='167',coloraxis='scan')
Zooming in on each dataset we see that they are about 1 hour long (you can also check the listobs output), and that each dataset has 2 or 3 observations of field 12. Each time, it's observed for about 25 seconds. We set solint='3600s' to get one solution per dataset.
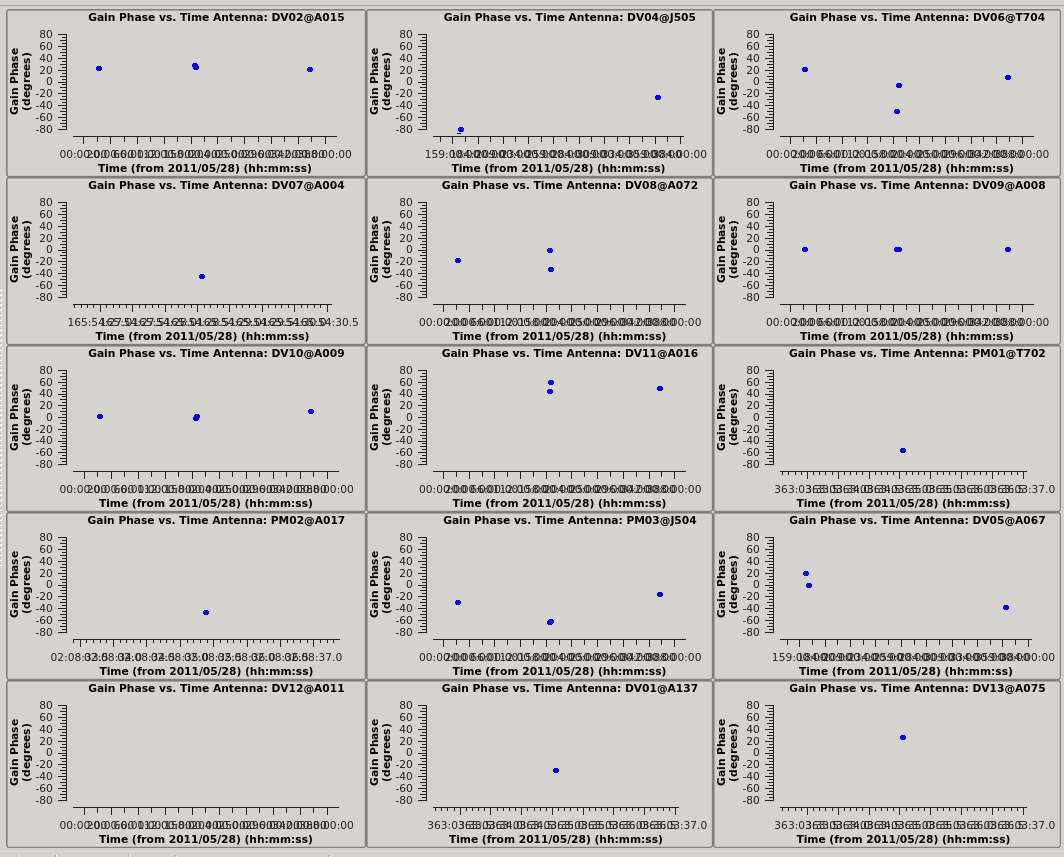
# In CASA
gaincal(vis='Antennae_North.cal.ms.contsub',caltable='north_self_1.pcal',
solint='3600s',combine='scan',gaintype='T',field='12',
refant='DV09',spw='0:80~92',minblperant=4,
calmode='p',minsnr=2)
Plot the calibration solutions.
# In CASA
plotcal(caltable='north_self_1.pcal',xaxis='time',yaxis='phase',
spw='',field='',antenna='',
iteration='antenna',subplot=531,plotrange=[0,0,-80,80],
figfile='north_pcal1_phase.png')
Apply the solutions to all fields and channels with applycal, which will overwrite the corrected data column:
# In CASA
applycal(vis='Antennae_North.cal.ms.contsub',field='',
gaintable=['north_self_1.pcal'],calwt=F)
Re-image the data with the selfcal applied. Remember that clean always uses the corrected data column if it exists:
# In CASA
os.system('rm -rf Antennae_North.CO3_2Line.Clean.pcal1*')
clean(vis='Antennae_North.cal.ms.contsub',
imagename='Antennae_North.CO3_2Line.Clean.pcal1',
spw='0',field='',phasecenter='12',
mode='velocity',outframe='LSRK',restfreq='345.79599GHz',
nchan=70,start='1200km/s',width='10km/s',
imagermode='mosaic',
imsize=500,cell='0.13arcsec',minpb=0.2,
interactive=T,mask='Antennae_North.CO3_2Line.Clean.mask',
weighting='briggs',robust=0.5,
niter=10000, threshold='5.0mJy')
After 600 iterations or so, inspect each channel to see if there is more emission that needs to be included in the mask. Remember to select the "all channels" toggle. Stop after 1200 or so iterations.
Compare the un-selfcal'ed and selfcal'ed data to determine if there has been improvement.
# In CASA
imview (raster=[{'file': 'Antennae_North.CO3_2Line.Clean.image',
'range': [-0.07,0.4]},
{'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image',
'range': [-0.07,0.4]}],
zoom={'channel': 43})
Then in the viewer, select the blink toggle in the "tapedeck", now the tapedeck buttons cycle between the two images at the selected channel. To change the channel toggle back to "normal", change the channel number, and then toggle back to "blink". To make the figure shown in 9.d, you can select the "p wrench" icon and change the number of panels in x to 2. You will need to drag your viewer window rather wide to get both panels to fit well. If you want to compare other channels, you need to toggle blink off, then use the tapedeck to move to the new channel of interest, and then turn blink back on to see both cubes again.
Note: while in "blink" mode, statistics will be for the final image in the stack. In "normal" mode, all open raster images will have their statistics reported.
What you should notice is that the peak flux density has increased substantially while the rms noise is about the same. This is reasonable for this self-calibration case because we have mostly adjusted relative position offsets between the datasets and not taken out any short-term phase variations which would reduce the rms.
Note that attempts to push the selfcal to shorter solint (for example to each scan of field=12) did not yield additional improvement.
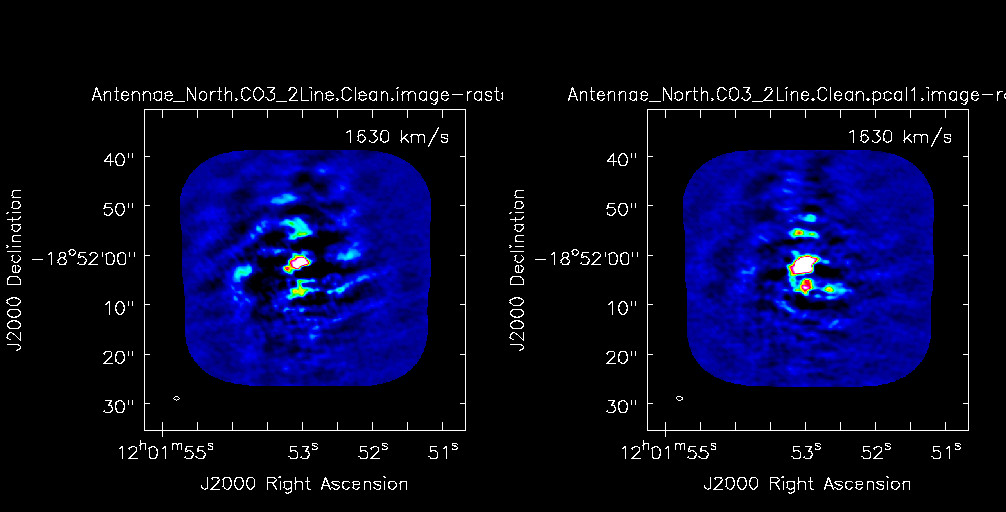
Southern Mosaic
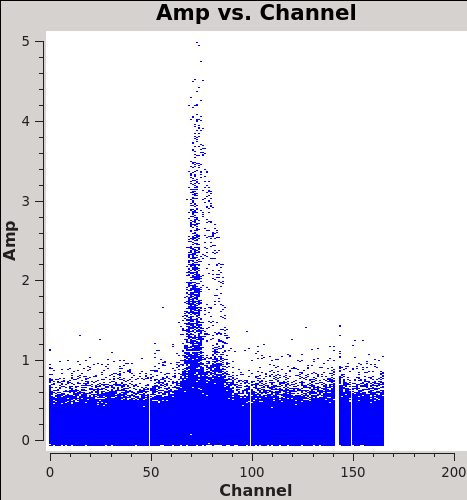
Now repeat the self-calibration process for the Southern mosaic. It is a bit tougher in this case to pick the best field. The most compact bright emission is toward field id 11 in the Overview. Adjusting for the renumbering of field ids after the final calibration split, this becomes field='7'.
From the following plots we can assess the brightest channels to pick and see that the timing in the 6 Southern datasets is similar to the North, with each lasting about 1 hour.
# In CASA
plotms(vis='Antennae_South.cal.ms.contsub',spw='',xaxis='channel',yaxis='amp',field='7',
avgtime='1e8',avgscan=T,coloraxis='field')
# In CASA
plotms(vis='Antennae_South.cal.ms.contsub',spw='',xaxis='time',yaxis='amp',field='7',
avgchannel='167',avgscan=T,coloraxis='scan')
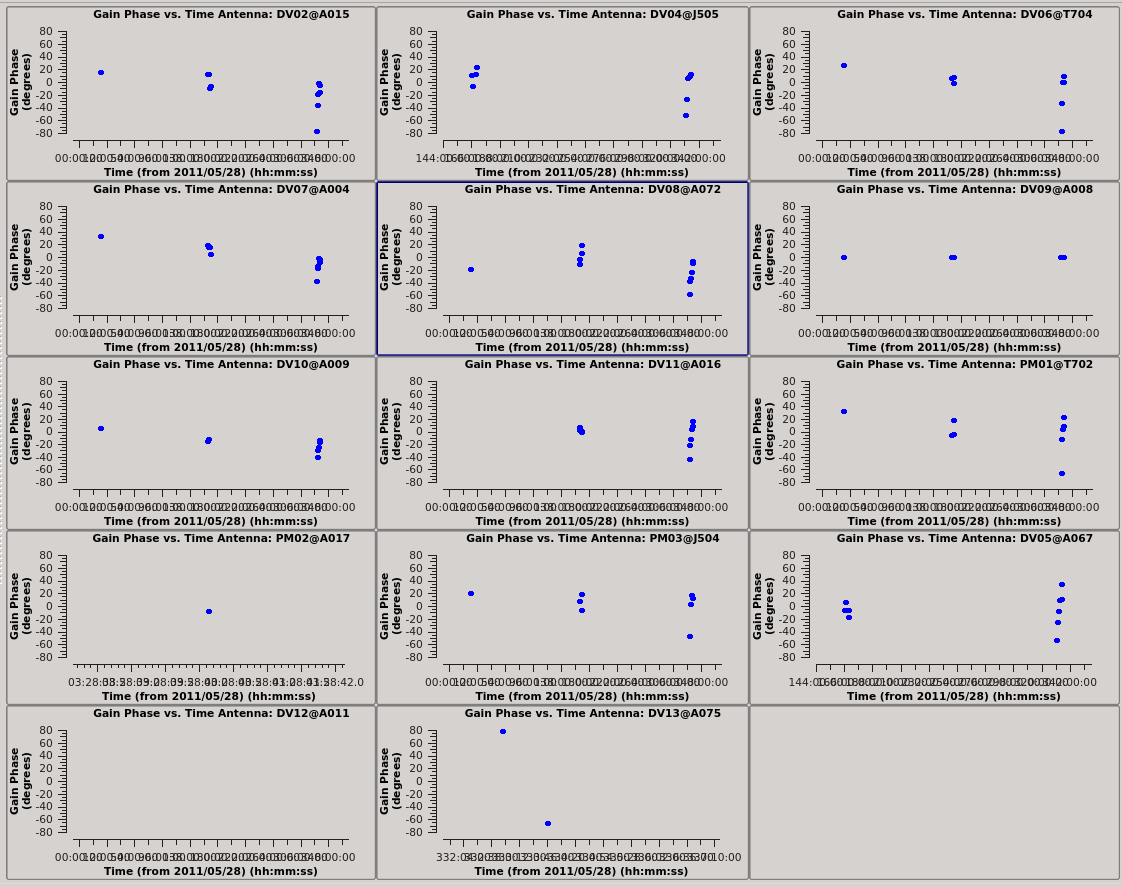
# In CASA
gaincal(vis='Antennae_South.cal.ms.contsub',caltable='south_self_1.pcal',
solint='3600s',combine='scan',gaintype='T',field='7',
refant='DV09',spw='0:69~80',minblperant=4,
calmode='p',minsnr=2)
# In CASA
plotcal(caltable='south_self_1.pcal',xaxis='time',yaxis='phase',
spw='',field='',antenna='',
iteration='antenna',subplot=531,plotrange=[0,0,-80,80],
figfile='south_pcal1_phase.png')
# In CASA
applycal(vis='Antennae_South.cal.ms.contsub',field='',
gaintable=['south_self_1.pcal'],calwt=F)
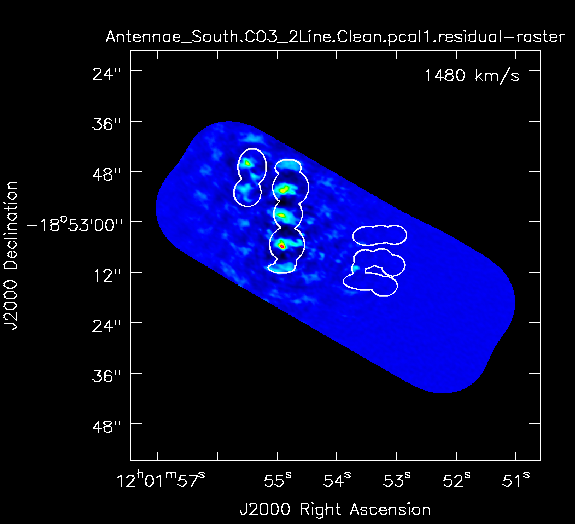
# In CASA
os.system('rm -rf Antennae_South.CO3_2Line.Clean.pcal1*')
clean(vis='Antennae_South.cal.ms.contsub',
imagename='Antennae_South.CO3_2Line.Clean.pcal1',
spw='0',field='',phasecenter='15',
mode='velocity',outframe='LSRK',restfreq='345.79599GHz',
nchan=70,start='1200km/s',width='10km/s',
imagermode='mosaic',
imsize=750,cell='0.13arcsec',minpb=0.2,
interactive=T,mask='Antennae_South.CO3_2Line.Clean.mask',
weighting='briggs',robust=0.5,
niter=10000, threshold='5.0mJy')
After 600 iterations or so, inspect each channel to see if there is more emission that needs to be included in the mask. Remember to select the "all channels" toggle. Stop after 2200 or so iterations.
As for the Northern mosaic, you can compare the before and after self-cal images:
# In CASA
imview (raster=[{'file': 'Antennae_South.CO3_2Line.Clean.image',
'range': [-0.05,0.2]},
{'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image',
'range': [-0.05,0.2]}],
zoom={'channel': 31})
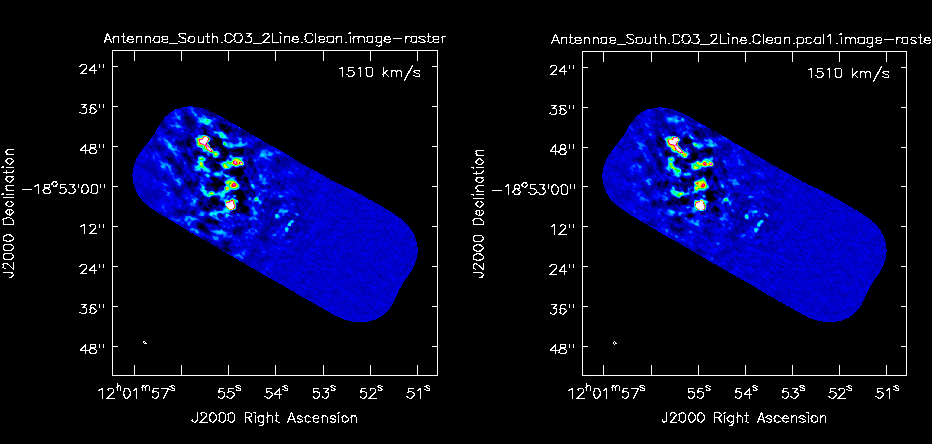
Image Analysis
Moment Maps
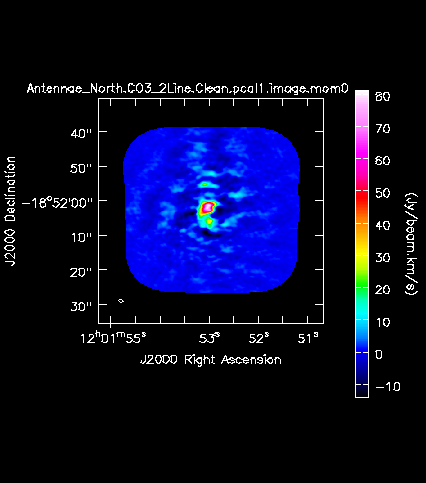
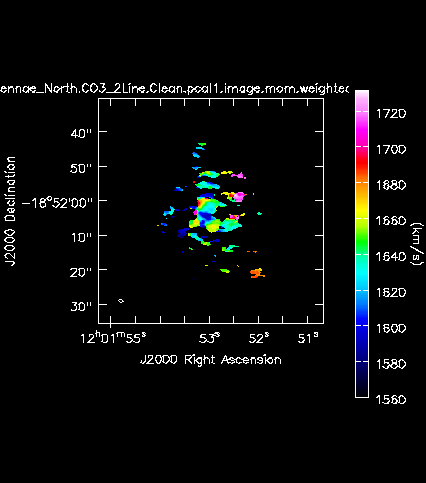
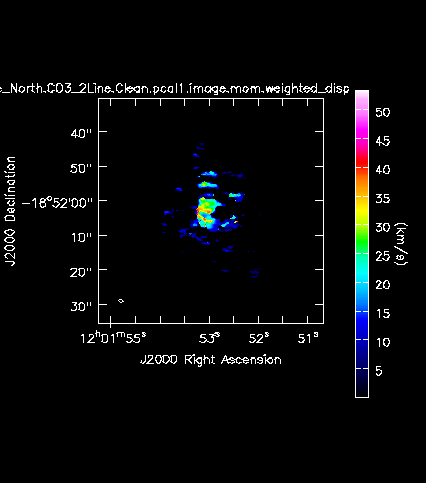
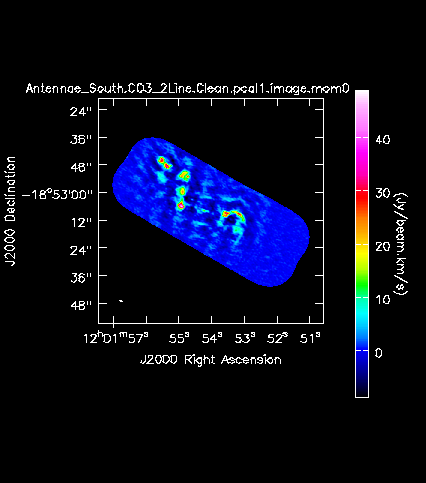
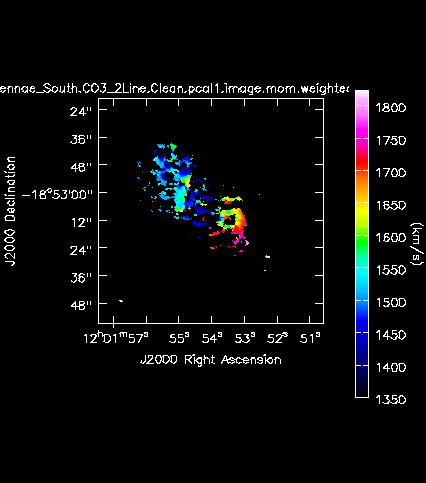
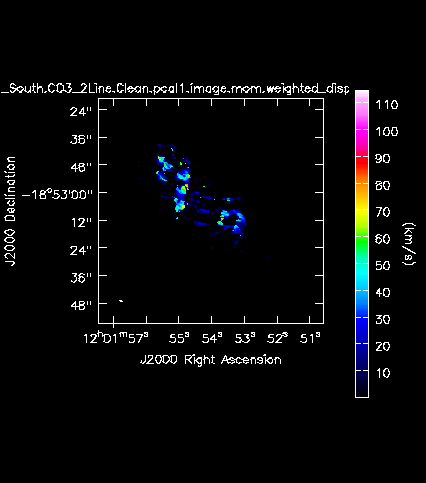
Next we will make moment maps for the CO(3-2) emission: Moment 0 is the integrated intensity; Moment 1 is the intensity weighted velocity field; and Moment 2 is the intensity weighted velocity dispersion.
Above we determined the rms noise levels for both the North and South mosaics in both a line-free and a line-bright channel. We want to limit the channel range of the moment calculations to those channels with significant emission. One good way to do this is to open the cube in the viewer overlaid with 3sigma contours, with sigma corresponding to the line-free rms.
# In CASA
imview(raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image',
'range': [-0.04,0.4]},
contour={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image',
'levels': [0.004],'unit': 5})
We find a channel range for significant emission of 33~63.
# In CASA
imview(raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image',
'range': [-0.04,0.4]},
contour={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image',
'levels': [0.0035],'unit': 5})
We find a channel range for significant emission of 12~64.
For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity.
# In CASA
immoments('Antennae_North.CO3_2Line.Clean.pcal1.image',
moments=[0],chans='33~63',
outfile='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0')
# In CASA
immoments('Antennae_South.CO3_2Line.Clean.pcal1.image',
moments=[0], chans='12~64',
outfile='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0')
For higher order moments it is very important to set a conservative flux threshold. Typically something like 3sigma, using sigma from a bright line channel works well. We do this with the mask parameter in the commands below. When making multiple moments, immoments uses the out parameter as a base and appends an appropriate ending.
# In CASA
immoments('Antennae_North.CO3_2Line.Clean.pcal1.image',
moments=[1,2], chans='33~63',
mask='Antennae_North.CO3_2Line.Clean.pcal1.image>0.018*3',
outfile='Antennae_North.CO3_2Line.Clean.pcal1.image.mom')
# In CASA
immoments('Antennae_South.CO3_2Line.Clean.pcal1.image',
moments=[1,2], chans='12~64',
mask=' Antennae_South.CO3_2Line.Clean.pcal1.image>0.009*3',
outfile='Antennae_South.CO3_2Line.Clean.pcal1.image.mom')
Next we can create the six moment map figures (11abc, 12abc) from these images with imview.
# In CASA
os.system('rm -f Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.png')
imview (raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image.mom0',
'colorwedge':T,'scaling': -0.5},
zoom=1,out='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.png')
os.system('rm -f Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png')
imview (raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord',
'colorwedge':T},
zoom=1,out='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png')
os.system('rm -f Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png')
imview (raster={'file': 'Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord',
'colorwedge':T},
zoom=1,out='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png')
os.system('rm -f Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.png')
imview (raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image.mom0',
'colorwedge':T,'scaling': -0.5},
zoom=1,out='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.png')
os.system('rm -f Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png')
imview (raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord',
'colorwedge':T},
zoom=1,out='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.png')
os.system('rm -f Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png')
imview (raster={'file': 'Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord',
'colorwedge':T},
zoom=1,out='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.png')
Exporting Images to Fits
If you want to analyze the data using another software package it is easy to convert from CASA format to FITS.
# In CASA
exportfits(imagename='Antennae_North.Cont.Clean.image',
fitsimage='Antennae_North.Cont.Clean.image.fits')
exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image',
fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.fits')
exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0',
fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.mom0.fits')
exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord',
fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.fits')
exportfits(imagename='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord',
fitsimage='Antennae_North.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.fits')
# In CASA
exportfits(imagename='Antennae_South.Cont.Clean.image',
fitsimage='Antennae_South.Cont.Clean.image.fits')
exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image',
fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.fits')
exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0',
fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.mom0.fits')
exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord',
fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_coord.fits')
exportfits(imagename='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord',
fitsimage='Antennae_South.CO3_2Line.Clean.pcal1.image.mom.weighted_dispersion_coord.fits')
Although "fits format" is supposed to be a standard, in fact most packages expect slightly different things from a fits image. If you are having difficulty, try setting velocity=T and/or dropstokes=T.
Comparison with previous SMA CO(3-2) data
We compare with SMA CO(3-2) data (Ueda, Iono, Petitpas et al., submitted to ApJ). Figure 13 shows a comparison plot between the moment 0 maps of ALMA and SMA data using the viewer. The fluxes, peak locations, and large scale structure are consistent. Both southern and northern components have been combined.
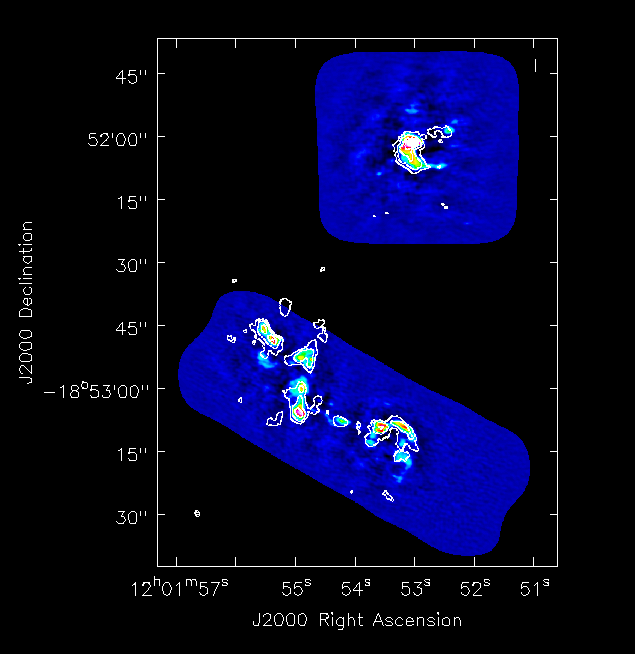
Last checked on CASA Version 3.3.0.