SMA CO Line Data 3.2: Difference between revisions
No edit summary |
|||
| (24 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
This script describes the data reduction of an SMA six-pointing mosaic of 12CO 2-1 emission in the infrared dark cloud G19.3+0.07. The CO line was placed in the USB. These data are from Brogan et al. in prep.; please do not use them for scientific purposes. This tutorial is based on the SMA CO Line Data script found in the [http://casa.nrao.edu/casascripts.shtml CASA Scripts and Data] page. | This script describes the data reduction of an SMA six-pointing mosaic of 12CO 2-1 emission in the infrared dark cloud G19.3+0.07. The CO line was placed in the USB. These data are from Brogan et al. in prep.; please do not use them for scientific purposes. This tutorial is based on the SMA CO Line Data script found in the [http://casa.nrao.edu/casascripts.shtml CASA Scripts and Data] page. | ||
The data were previously filled in [http://www.atnf.csiro.au/computing/software/miriad/ Miriad]. A Tsys correction was applied, and the data were then written out in uvfits format. In this tutorial, CASA is used to read the uvfits files, and flag, calibrate, continuum subtract and image the data | The data were previously filled in [http://www.atnf.csiro.au/computing/software/miriad/ Miriad]. A Tsys correction was applied, and the data were then written out in uvfits format. In this tutorial, CASA is used to read the uvfits files, and flag, calibrate, continuum subtract and image the data. | ||
You will need the tarball of the uvfits data, which can be downloaded [http://casa.nrao.edu/Data/SMA/spw_fits_files.tar here]. | You will need the tarball of the uvfits data, which can be downloaded [http://casa.nrao.edu/Data/SMA/spw_fits_files.tar here]. | ||
<pre style="background-color: #FFFF00;"> | <pre style="background-color: #FFFF00;"> | ||
Where possible the new plotter plotms has been used, as the old plotter, plotxy, can be slow for | |||
datasets with many spw such as SMA data. We have continued to use plotxy in some cases where multipanel | |||
plots are useful, as plotms can not yet display multipanel plots. In other places, we use plotms with | |||
the iteraxis parameter set. This will cause plotms to create multiple plots over the set iteraxis | |||
parameter, which you can view by pressing the arrow keys on the plotms display. Plotxy will eventually | |||
be phased out, and this tutorial will be updated. In the meantime, this tutorial will work with | be phased out, and this tutorial will be updated. In the meantime, this tutorial will work with | ||
CASA Version 3. | CASA Version 3.2.0. | ||
</pre> | </pre> | ||
| Line 166: | Line 170: | ||
</div> | </div> | ||
[[File: | [[File:ampvsfreq_clr.jpg|thumb| Amplitude vs. frequency for the bandpass calibrator 3c454.3.]] | ||
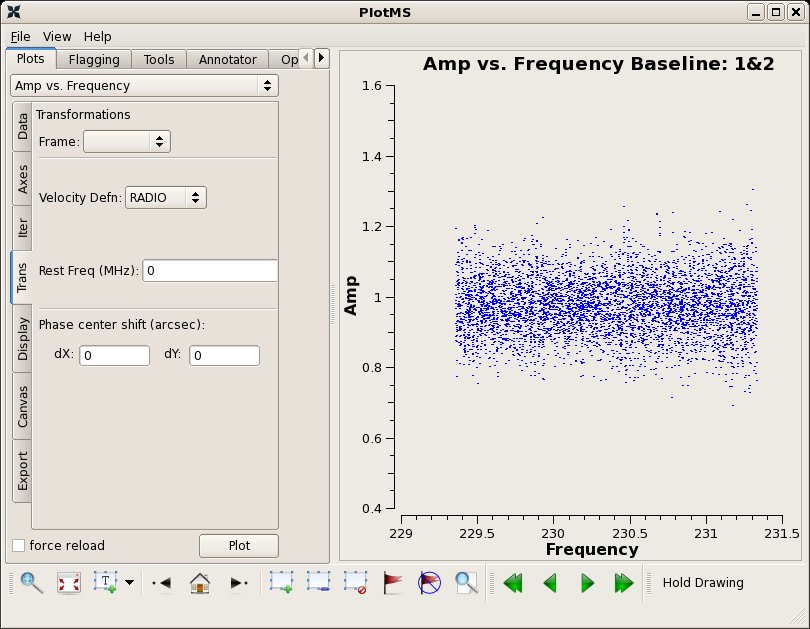
Make an inital amplitude vs. frequency plot of the bandpass calibrator 3c454.3, averaging the data together in time. Here, we colorize the points by spw in the {{plotms}} call using the '''coloraxis''' parameter. You can also do this under the 'Display' tab in the plotms window. | Make an inital amplitude vs. frequency plot of the bandpass calibrator 3c454.3, averaging the data together in time. Here, we colorize the points by spw in the {{plotms}} call using the '''coloraxis''' parameter. You can also do this under the 'Display' tab in the plotms window. | ||
| Line 185: | Line 189: | ||
</source> | </source> | ||
[[File: | [[File:ampvstime_clr.jpg|thumb| Amplitude vs. Time before flagging first integrations.]] | ||
On many baselines the first integration of every scan is low. This can be seen with {{plotms}}. | On many baselines the first integration of every scan is low. This can be seen with {{plotms}}. | ||
| Line 196: | Line 200: | ||
Here, we have colored the points by field to see that the low amplitudes are found in the first integrations of many scans. These can also be flagged with {{flagdata}}. The integration time of these data is 30.9s. Note that more recent SMA data rarely show this problem. | Here, we have colored the points by field to see that the low amplitudes are found in the first integrations of many scans. These can also be flagged with {{flagdata}}. The integration time of these data is 30.9s. Note that more recent SMA data rarely show this problem. | ||
[[File: | [[File:plotxy-ants.png|thumb| Antenna positions for these data.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 218: | Line 222: | ||
== Bandpass Calibration == | == Bandpass Calibration == | ||
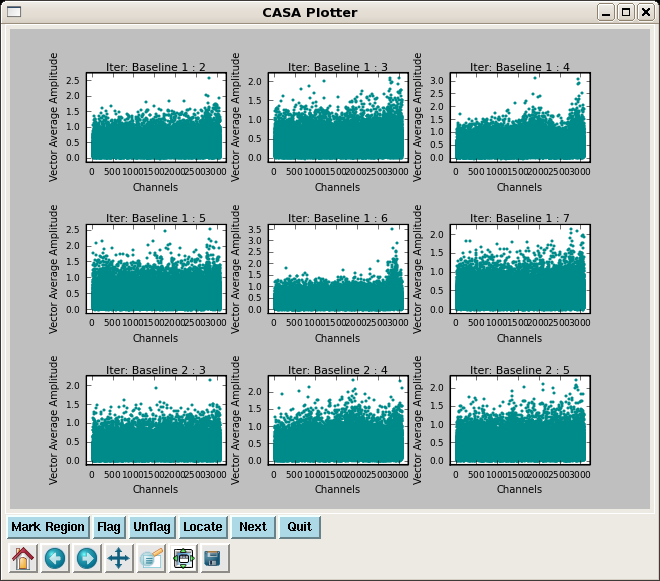
Next plot phase vs. channel. Here, we use plotms to iterate over the SMA baselines. While multiple subplots are not yet operational within plotms, you can view the individual phase vs. channel plots using the arrow buttons at the bottom right of the {{plotms}} window. By setting the '''xaxis''' parameter to 'frequency', we can see how the spectral windows overlap. Note the large discrepancy in phase for spw=4-7 on some baselines - this will be important later. | |||
[[File:phaseVsfreqBaseline13.jpg|thumb| Phase vs. channel for the bandpass calibrator 3c454.3 and baseline 1-3.]] | |||
[[File:phaseVsfreqBaseline56.jpg|thumb| Phase vs. channel for the bandpass calibrator 3c454.3 and baseline 5-6.]] | |||
<source lang="python"> | <source lang="python"> | ||
| Line 265: | Line 270: | ||
[[File:plotxy-pha_vs_time.png|thumb| Phase vs. time for the bandpass calibrator 3c454.3.]] | [[File:plotxy-pha_vs_time.png|thumb| Phase vs. time for the bandpass calibrator 3c454.3.]] | ||
Here we plot phase vs. time for the bandpass calibrator 3c454.3 | Here we plot phase vs. time for the bandpass calibrator 3c454.3 using {{plotxy}}: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
| Line 371: | Line 369: | ||
Although in previous steps solutions were applied "on-the-fly", the data were not actually changed. Running {{applycal}} will fill the corrected data column in the ms. Again be careful with specification of '''gaintable''', and keeping the same order in '''spwmap''' and '''gainfield'''. This {{applycal}} is not necessary for later steps, but allows plotting of data calibration to this point. | Although in previous steps solutions were applied "on-the-fly", the data were not actually changed. Running {{applycal}} will fill the corrected data column in the ms. Again be careful with specification of '''gaintable''', and keeping the same order in '''spwmap''' and '''gainfield'''. This {{applycal}} is not necessary for later steps, but allows plotting of data calibration to this point. | ||
[[File: | [[File:applycal-plotms-Baseline34.jpg|thumb| Gain-corrected 3c454.3 data. Notice the few residual baseline-based problems.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 421: | Line 419: | ||
Now we can do a new {{applycal}} including ''only'' the second blcal2 solution. Note that {{applycal}} overwrites the corrected column, so there is no need to {{clearcal}}. Like the antenna based bandpass the {{blcal}} solution is independent for each spw, so we set '''spwmap = []'''. | Now we can do a new {{applycal}} including ''only'' the second blcal2 solution. Note that {{applycal}} overwrites the corrected column, so there is no need to {{clearcal}}. Like the antenna based bandpass the {{blcal}} solution is independent for each spw, so we set '''spwmap = []'''. | ||
[[File: | [[File:bandpass-finalfixed.jpg|thumb| Gain-corrected 3c454.3 data, with residual problems fixed.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 478: | Line 476: | ||
</source> | </source> | ||
[[File: | [[File:plotxyAmpvsTimeFld1.jpg|thumb| Corrected amplitude vs. time for 1743-038.]] | ||
[[File:plotxy-pha_vs_time-corr1.png|thumb| Corrected phase vs. time for 1743-038.]] | [[File:plotxy-pha_vs_time-corr1.png|thumb| Corrected phase vs. time for 1743-038.]] | ||
Have a look at the corrected data for both calibrators in phase and amplitude vs. time. | Have a look at the corrected data for both calibrators in phase and amplitude vs. time. We can't color by field in {{plotxy}}, unfortunately, so this is easiest to do separately. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
# First, amplitude vs. time for field 1: | # First, amplitude vs. time for field 1: | ||
plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",datacolumn="corrected", | plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",datacolumn="corrected", | ||
iteration="baseline", antenna="",spw="0~23",field="1", | iteration="baseline", antenna="",spw="0~23",field="1", | ||
| Line 609: | Line 606: | ||
The final application of calibration for each source is done independently below. Flux densities from SMA flux monitoring are provided as checks of the absolute flux calibration. | The final application of calibration for each source is done independently below. Flux densities from SMA flux monitoring are provided as checks of the absolute flux calibration. | ||
[[File: | [[File:FinalAmpvsTimeField9.jpg|thumb| Corrected amplitude vs. time for 3C454.3.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 627: | Line 624: | ||
</pre> | </pre> | ||
[[File: | [[File:FinalAmpvsTimeField1.jpg|thumb| Corrected amplitude vs. time for 1743-038.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 645: | Line 642: | ||
</pre> | </pre> | ||
[[File: | [[File:FinalAmpvsTimeField6.jpg|thumb| Corrected amplitude vs. time for 1908-201.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 664: | Line 661: | ||
</pre> | </pre> | ||
[[File: | [[File:UranusAmpvsUVdist.jpg|thumb| Corrected amplitude vs. uvdistance for Uranus.]] | ||
Since Uranus is resolved we apply the solutions from 3C454.3 to achieve at least some reduction in instrumental gain variations. | Since Uranus is resolved we apply the solutions from 3C454.3 to achieve at least some reduction in instrumental gain variations. | ||
| Line 720: | Line 717: | ||
</div> | </div> | ||
[[File: | [[File:sourceAmpvsFreq.jpg|thumb| Amplitude vs. frequency for the target data.]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 732: | Line 729: | ||
avgchannel='3072',avgspw=True) | avgchannel='3072',avgspw=True) | ||
</source> | </source> | ||
[[File:sourceElvsTime.jpg|thumb| Elevation vs. Time for the target data.]] | |||
Data at times > 11:04:00 show lots of scatter: the elevation of these data are very low and are worth flagging. One can {{plotms}} with '''xaxis="time"''', '''yaxis="elevation"''' to see this. | Data at times > 11:04:00 show lots of scatter: the elevation of these data are very low and are worth flagging. One can {{plotms}} with '''xaxis="time"''', '''yaxis="elevation"''' to see this. | ||
| Line 739: | Line 738: | ||
plotms(vis='g19_d2usb_targets.ms',xaxis="time",yaxis="elevation",field='') | plotms(vis='g19_d2usb_targets.ms',xaxis="time",yaxis="elevation",field='') | ||
</source> | </source> | ||
<source lang="python"> | <source lang="python"> | ||
| Line 746: | Line 744: | ||
</source> | </source> | ||
[[File: | [[File:sourceAmpvsVel.jpg|thumb| Amplitude vs. velocity for the target data.]] | ||
Note that because the Doppler tracked frequency was 13CO (+ 10GHz in the USB), the velocity diplayed | Note that because the Doppler tracked frequency was 13CO (+ 10GHz in the USB), the velocity diplayed in {{plotms}} will not be correct unless you set the rest frequency to the line of interest. You can do this in the {{plotms}} call, or in the '''Trans''' tab in the {{plotms}} display: | ||
CO(2-1) restfreq=230.53797 GHz | CO(2-1) restfreq=230.53797 GHz | ||
| Line 755: | Line 753: | ||
# In CASA | # In CASA | ||
clearstat() | clearstat() | ||
plotms(vis="g19_d2usb_targets.ms",xaxis="velocity",yaxis="amp", | |||
ydatacolumn="data",selectdata=True, | |||
spw="0~23",field="0", | |||
averagedata=True,avgtime='99999',avgscan=True, | |||
transform=True,restfreq="230.53797GHz") | |||
</source> | </source> | ||
| Line 769: | Line 766: | ||
== Continuum imaging and subtraction == | == Continuum imaging and subtraction == | ||
Next we want to make a continuum-free line dataset. We use field 0 to pick the line free spw | Next we want to make a continuum-free line dataset. We use field 0 to pick the line free spw, and can see that the line is confined to spws 13~15 by colorizing by spw: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
clearstat() | clearstat() | ||
plotms(vis="g19_d2usb_targets.ms",xaxis="freq",yaxis="amp", | |||
ydatacolumn="data",selectdata=True,coloraxis="spw", | |||
spw="0~23",field="0",averagedata=True,avgtime='99999') | |||
</source> | </source> | ||
Copy the original ms in case something goes wrong. Currently {{uvcontsub2}} will change the input model and corrected columns, so it's best to make a back up. This behavior will change in future. | Copy the original ms in case something goes wrong. Currently {{uvcontsub2}} will change the input model and corrected columns, so it's best to make a back up. This behavior will change in future. | ||
| Line 796: | Line 788: | ||
# In CASA | # In CASA | ||
uvcontsub2(vis='g19_d2usb_targets_line.ms',fitspw='0~12,16~23', | uvcontsub2(vis='g19_d2usb_targets_line.ms',fitspw='0~12,16~23', | ||
solint='int',combine='spw' | solint='int',combine='spw',spw='0~23', | ||
fitorder=1) | |||
</source> | </source> | ||
The directory g19_d2usb_targets_line.ms. | The directory g19_d2usb_targets_line.ms.contsub/ contains the continuum-subtracted line data. We will make a separate dataset, g19_d2usb_targets_line.ms.cont, using the {{split}} task for imaging. Because the continuum model subtracted from the line dataset by {{uvcontsub2}} is necessarily a smoothed fit, images made with it are liable to have their field of view reduced in some strange way. Images of the continuum should be made by simply excluding the line channels, as we show here. | ||
<source lang="python"> | |||
# In CASA | |||
split(vis='g19_d2usb_targets.ms',outputvis='g19_d2usb_targets_line.ms.cont', | |||
datacolumn='data',field='',spw='0~12,16~23') | |||
</source> | |||
Next we image the continuum data: | Next we image the continuum data: | ||
[[File:viewer-contdata.jpg|thumb| Viewer display of the continuum data.]] | |||
<div style="background-color: #dddddd;"> | <div style="background-color: #dddddd;"> | ||
| Line 817: | Line 818: | ||
phasecenter="J2000 18h25m56.09 -12d04m28.20", | phasecenter="J2000 18h25m56.09 -12d04m28.20", | ||
pbcor=F,minpb=0.2) | pbcor=F,minpb=0.2) | ||
viewer('g19_d2usb_cont.image') | |||
</source> | </source> | ||
Here you can see one stronger continuum source to the NE (18:25:58.5 -12:03:58.9) and one weak one to the SW (18:25:52, -12:04:53). The weak one is only visible after one round of cleaning. Because the mosaic is sparse you should make a clean mask to get the best cleaned image. | Here you can see one stronger continuum source to the NE (18:25:58.5 -12:03:58.9) and one weak one to the SW (18:25:52, -12:04:53). The weak one is only visible after one round of cleaning. Because the mosaic is sparse you should make a clean mask to get the best cleaned image. | ||
[[File: | [[File:viewer-contsubdata.jpg|thumb| Viewer display of the continuum-subtracted data, excluding line spws.]] | ||
Make a dirty image of the continuum subtracted data, excluding the spws which contain line emission. You should see nothing but noise. | Make a dirty image of the continuum subtracted data, excluding the spws which contain line emission. You should see nothing but noise. | ||
| Line 846: | Line 849: | ||
== Spectral line imaging == | == Spectral line imaging == | ||
[[File: | [[File:viewer-linedata17kms.jpg|thumb| Viewer display of the {{clean}}-produced image at the 17 km/s channel.]] | ||
This image selects a channel range suitable to image the 12CO(2-1) line. Notice the use of the '''restfreq''' parameter to get the velocity scale correct. | This image selects a channel range suitable to image the 12CO(2-1) line. Notice the use of the '''restfreq''' parameter to get the velocity scale correct. | ||
| Line 865: | Line 868: | ||
</source> | </source> | ||
[[File: | [[File:viewer-linemom0.jpg|thumb| Viewer display of the moment 0 map.]] | ||
[[File: | [[File:viewer-linemom1.jpg|thumb| Viewer display of the moment 1 map.]] | ||
It is essential to run with '''interactive=T''' and make a clean mask. A clean mask is available in the original data directory spw_fits_files. To use the pre-made mask, run: | It is essential to run with '''interactive=T''' and make a clean mask. A clean mask is available in the original data directory spw_fits_files. To use the pre-made mask, run: | ||
| Line 919: | Line 922: | ||
{{Checked 3.2.0}} | {{Checked 3.2.0}} | ||
--[[User:Rfriesen|Rfriesen]] 10:25, 5 May 2011 (PDT) | |||
Latest revision as of 17:25, 5 May 2011
This script describes the data reduction of an SMA six-pointing mosaic of 12CO 2-1 emission in the infrared dark cloud G19.3+0.07. The CO line was placed in the USB. These data are from Brogan et al. in prep.; please do not use them for scientific purposes. This tutorial is based on the SMA CO Line Data script found in the CASA Scripts and Data page.
The data were previously filled in Miriad. A Tsys correction was applied, and the data were then written out in uvfits format. In this tutorial, CASA is used to read the uvfits files, and flag, calibrate, continuum subtract and image the data.
You will need the tarball of the uvfits data, which can be downloaded here.
Where possible the new plotter plotms has been used, as the old plotter, plotxy, can be slow for datasets with many spw such as SMA data. We have continued to use plotxy in some cases where multipanel plots are useful, as plotms can not yet display multipanel plots. In other places, we use plotms with the iteraxis parameter set. This will cause plotms to create multiple plots over the set iteraxis parameter, which you can view by pressing the arrow keys on the plotms display. Plotxy will eventually be phased out, and this tutorial will be updated. In the meantime, this tutorial will work with CASA Version 3.2.0.
Creating the measurement set from uvfits data
First, remove any files from previous run-throughs and unpack the tarball:
# In CASA
# Remove any files from previous run-throughs
os.system('rm -rf spw_fits.files')
os.system('tar xvf spw_fits_files.tar')
os.system('rm -rf g19_d2usb*')
Read in each uvfits file using the importuvfits filler (you need to set the upper range of the for loop to one more than you actually have, to satisfy ipython peculiarity). We create a list of the uvfits filenames in myfiles that will be used to create a single measurement set (ms).
# In CASA
os.system('rm -rf spw_ms_files')
os.system('mkdir spw_ms_files')
myfiles = []
for i in range(1,25):
msfile = "spw_ms_files/g19_d2usb.spw"+str(i)+".ms"
importuvfits(fitsfile="spw_fits_files/g19_d2usb.spw"+str(i),
vis=msfile)
myfiles.append(msfile)
Using concat, create a single ms from the filenames listed in myfiles:
# In CASA
concat(vis=myfiles,concatvis='g19_d2usb.ms',timesort=True)
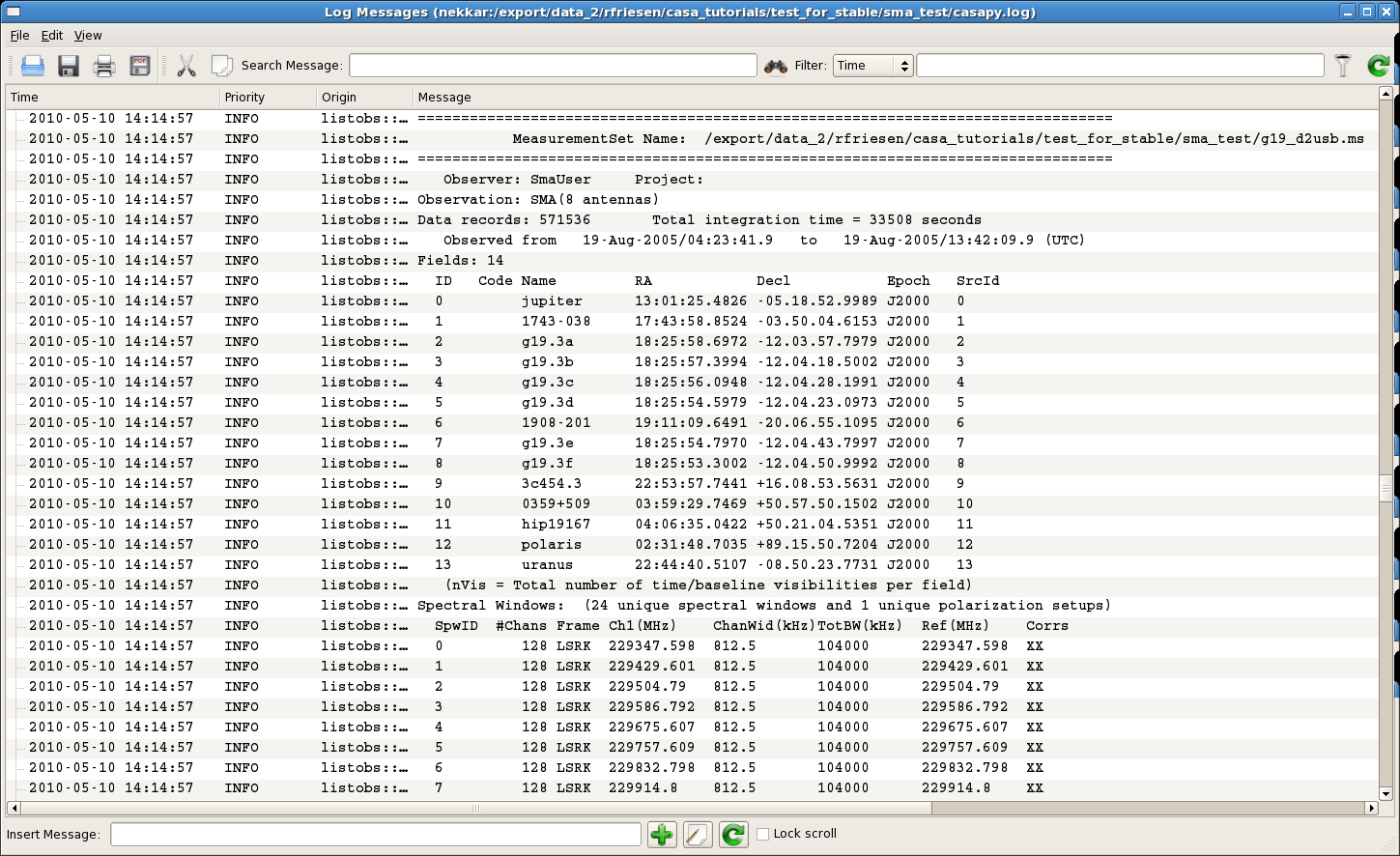
Finally, initialize the scratch columns using clearcal and use listobs to look at the contents of the measurement set (which appear in the CASA Log Messages window):
# In CASA
clearcal(vis='g19_d2usb.ms')
listobs(vis='g19_d2usb.ms',verbose=False)
##########################################
##### Begin Task: listobs #####
listobs::::casa
================================================================================
MeasurementSet Name: /export/data_2/rfriesen/casa_tutorials/test_for_stable/sma_co/sma_test/g19_d2usb.ms MS Version 2
================================================================================
Observer: SmaUser Project:
Observation: SMA(8 antennas)
Data records: 571536 Total integration time = 33508 seconds
Observed from 19-Aug-2005/04:23:41.9 to 19-Aug-2005/13:42:09.9 (UTC)
Fields: 14
ID Code Name RA Decl Epoch SrcId
0 jupiter 13:01:25.48 -05.18.53.00 J2000 0
1 1743-038 17:43:58.85 -03.50.04.62 J2000 1
2 g19.3a 18:25:58.70 -12.03.57.80 J2000 2
3 g19.3b 18:25:57.40 -12.04.18.50 J2000 3
4 g19.3c 18:25:56.09 -12.04.28.20 J2000 4
5 g19.3d 18:25:54.60 -12.04.23.10 J2000 5
6 1908-201 19:11:09.65 -20.06.55.11 J2000 6
7 g19.3e 18:25:54.80 -12.04.43.80 J2000 7
8 g19.3f 18:25:53.30 -12.04.51.00 J2000 8
9 3c454.3 22:53:57.74 +16.08.53.56 J2000 9
10 0359+509 03:59:29.75 +50.57.50.15 J2000 10
11 hip19167 04:06:35.04 +50.21.04.54 J2000 11
12 polaris 02:31:48.70 +89.15.50.72 J2000 12
13 uranus 22:44:40.51 -08.50.23.77 J2000 13
(nVis = Total number of time/baseline visibilities per field)
Spectral Windows:(24 unique spectral windows and 1 unique polarization setups)
SpwID #Chans Frame Ch1(MHz) ChanWid(kHz)TotBW(kHz) Ref(MHz) Corrs
0 128 LSRK 229347.598 812.5 104000 229347.598 XX
1 128 LSRK 229429.601 812.5 104000 229429.601 XX
2 128 LSRK 229504.79 812.5 104000 229504.79 XX
3 128 LSRK 229586.792 812.5 104000 229586.792 XX
4 128 LSRK 229675.607 812.5 104000 229675.607 XX
5 128 LSRK 229757.609 812.5 104000 229757.609 XX
6 128 LSRK 229832.798 812.5 104000 229832.798 XX
7 128 LSRK 229914.8 812.5 104000 229914.8 XX
8 128 LSRK 230003.615 812.5 104000 230003.615 XX
9 128 LSRK 230085.617 812.5 104000 230085.617 XX
10 128 LSRK 230160.806 812.5 104000 230160.806 XX
11 128 LSRK 230242.808 812.5 104000 230242.808 XX
12 128 LSRK 230331.623 812.5 104000 230331.623 XX
13 128 LSRK 230413.625 812.5 104000 230413.625 XX
14 128 LSRK 230488.814 812.5 104000 230488.814 XX
15 128 LSRK 230570.817 812.5 104000 230570.817 XX
16 128 LSRK 230659.631 812.5 104000 230659.631 XX
17 128 LSRK 230741.634 812.5 104000 230741.634 XX
18 128 LSRK 230816.823 812.5 104000 230816.823 XX
19 128 LSRK 230898.825 812.5 104000 230898.825 XX
20 128 LSRK 230987.639 812.5 104000 230987.639 XX
21 128 LSRK 231069.641 812.5 104000 231069.641 XX
22 128 LSRK 231144.831 812.5 104000 231144.831 XX
23 128 LSRK 231226.833 812.5 104000 231226.833 XX
The sources contained in this measurement set are:
Source Name Description ID ==================================================================== 3c454.3 Bandpass and flux calibrator 9 1743-038 Gain calibrator 1 1908-201 Gain calibrator 6 Uranus Flux calibrator 13 g19.7 Science target (6-pointing mosaic) 2,3,4,5,7,8 ====================================================================
Other sources in the ms were not part of this program.
Initial inspection and flagging
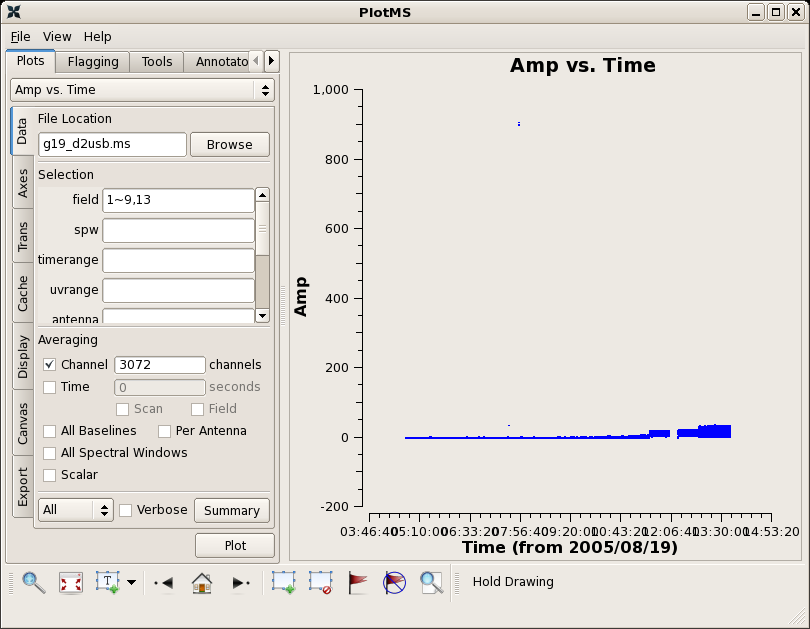
Use plotms to inspect the data.
# In CASA
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",field='1~9,13',
avgchannel='3072')
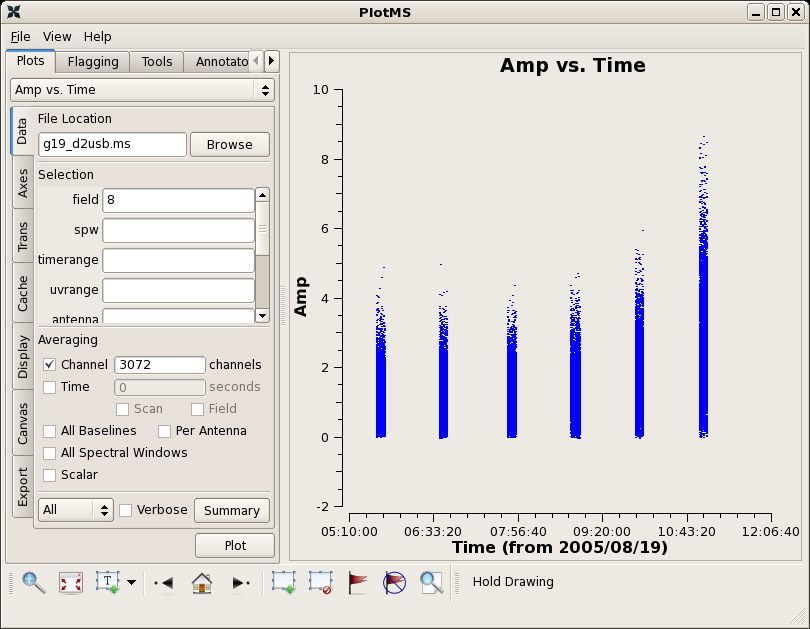
There are a few bad data points in this ms. You can use the locate features in plotms to find the bad data points on field 8 and antenna ids 1-2 and 0-2. Flagging can be done in plotms, or in a script using the flagdata command. Here we will do all flagging within the script.
Tip: locate gives antenna ids, but flagdata looks first at the name column. Since SMA antenna names are numbers, this can be confusing. The corresponding names are antennas 2-3 and 1-3, which you can check with listobs.
# In CASA
flagdata(vis='g19_d2usb.ms',mode='manualflag',field='8',spw='20~23',
timerange='07:52:45~07:52:47',antenna='2&3;1&3')
Plotting again shows that the bad data are gone:
# In CASA
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",field='8',
avgchannel='3072')
Tip: to confirm the data were flagged in plotms, you need to change selection in plot (for example change field and then change back), and then change back to original to clear the cache and force a re-read of the data.
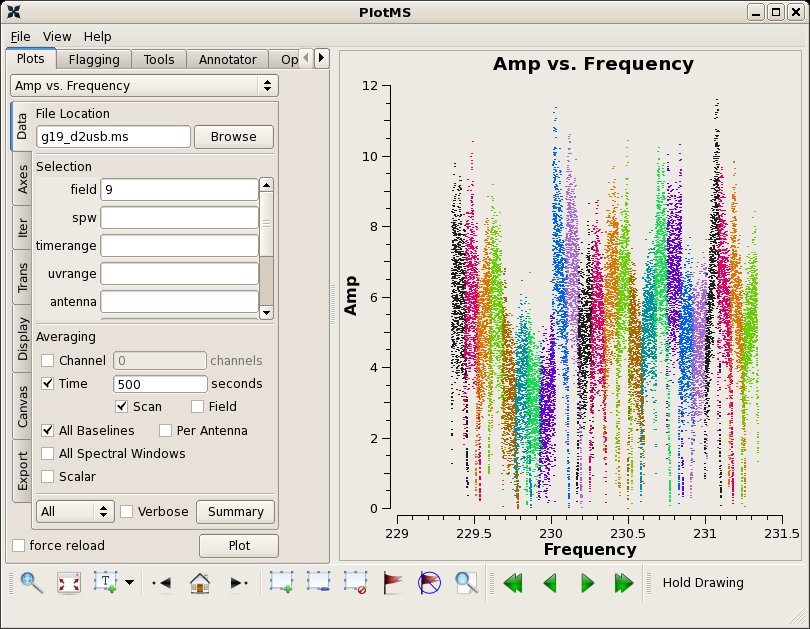
Make an inital amplitude vs. frequency plot of the bandpass calibrator 3c454.3, averaging the data together in time. Here, we colorize the points by spw in the plotms call using the coloraxis parameter. You can also do this under the 'Display' tab in the plotms window.
# In CASA
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="freq",yaxis="amp",field="9",
averagedata=True,avgscan=True,avgtime='500',avgbaseline=True,coloraxis="spw")
As you can see, SMA spw overlap by a fair amount. Here we will flag the first seven channels on either side of each spw to aid calibration:
# In CASA
default('flagdata')
flagdata(vis='g19_d2usb.ms', mode='manualflag',spw='0~23:0~6;121~127')
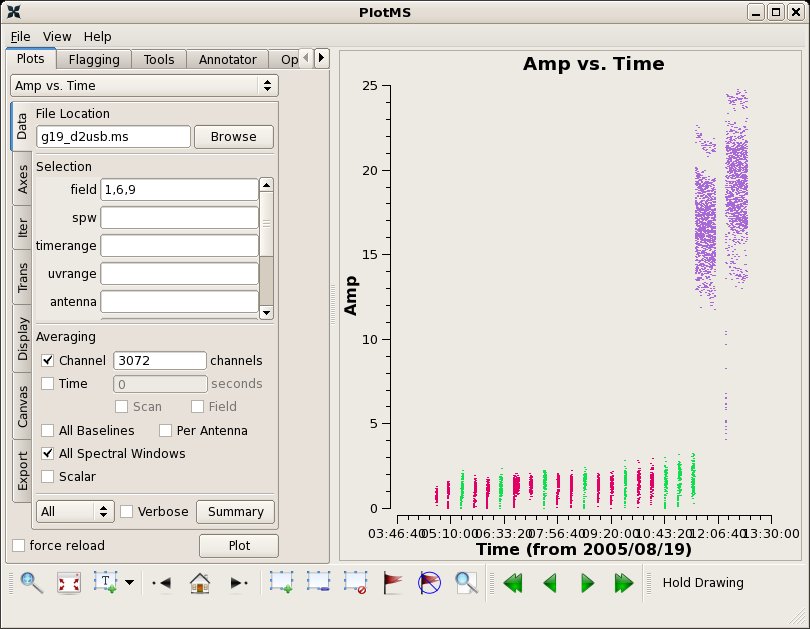
On many baselines the first integration of every scan is low. This can be seen with plotms.
# In CASA
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",
field='1,6,9',avgchannel='3072',avgspw=T,coloraxis="field")
Here, we have colored the points by field to see that the low amplitudes are found in the first integrations of many scans. These can also be flagged with flagdata. The integration time of these data is 30.9s. Note that more recent SMA data rarely show this problem.
# In CASA
flagdata(vis="g19_d2usb.ms",mode="quack",field='',
spw="0~23",quackinterval=40.0)
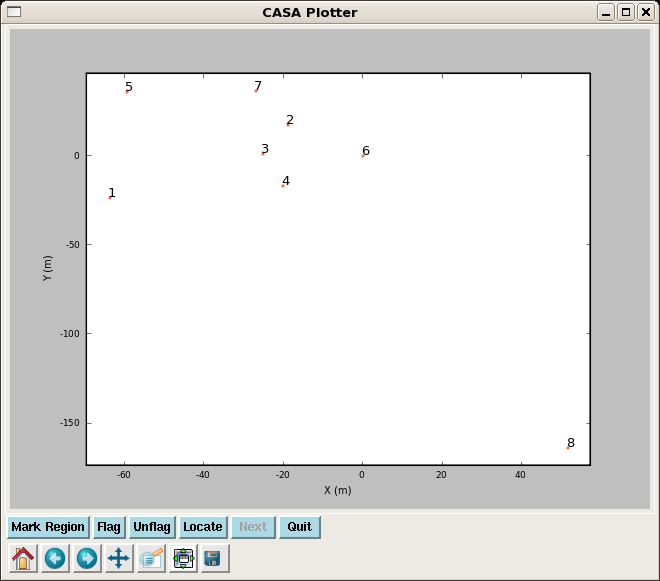
We can use plotxy to plot the antenna positions. For these observations, antenna 8 was in the barn for repair.
# In CASA
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="x")
Tip: after you run plotxy, run clearstat() before trying to run plotms or you will get a table lock.
Bandpass Calibration
Next plot phase vs. channel. Here, we use plotms to iterate over the SMA baselines. While multiple subplots are not yet operational within plotms, you can view the individual phase vs. channel plots using the arrow buttons at the bottom right of the plotms window. By setting the xaxis parameter to 'frequency', we can see how the spectral windows overlap. Note the large discrepancy in phase for spw=4-7 on some baselines - this will be important later.
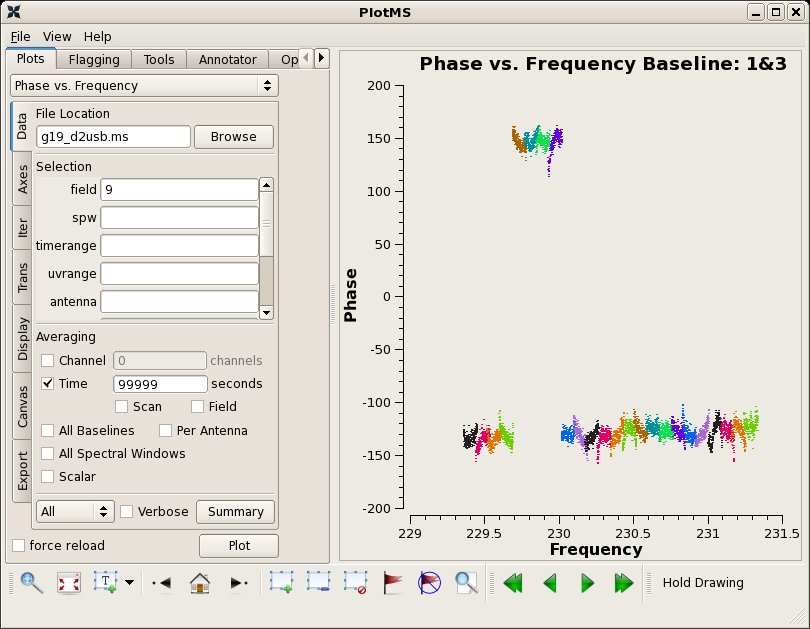
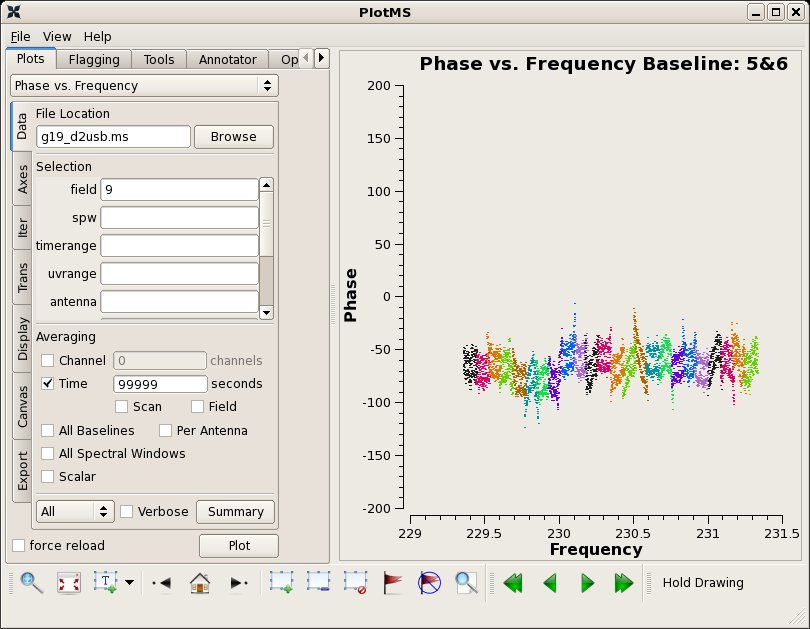
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="frequency",yaxis="phase",
ydatacolumn="data",field="9",iteraxis="baseline",
averagedata=True,avgtime="99999",coloraxis="spw")
We can also use the slower plotxy to show multiple subplots in one panel. We set the plotxy parameter crossscan=T because there are two scans on 3c454.3.
# In CASA
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="channel",yaxis="phase",datacolumn="data",
iteration="baseline",
antenna="",spw="",field="9",
averagemode="vector",width="1",timebin="all",
crossscans=T,crossbls=False,stackspw=F,
subplot=331,plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
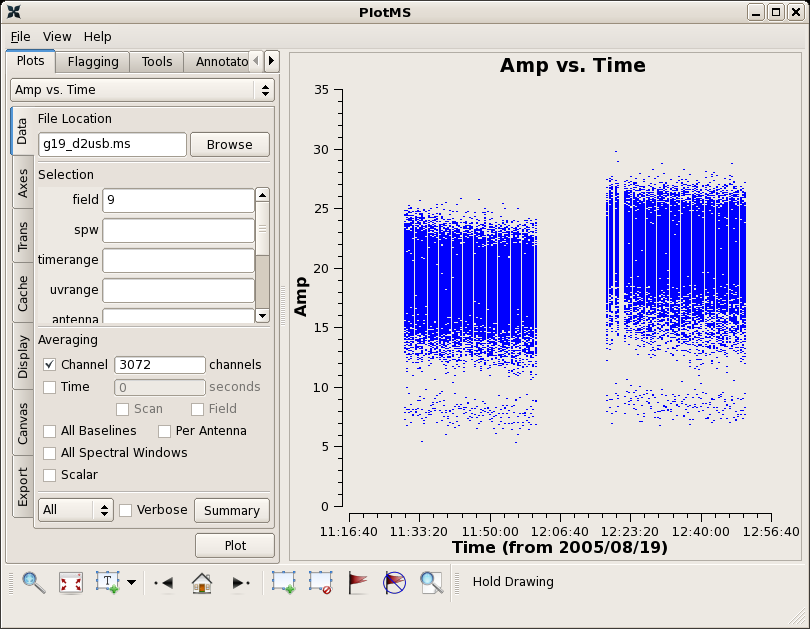
Now plot amplitude and phase as a function of time.
# In CASA
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",field='9',
avgchannel='3072')
The gap in the 3C454.3 observation is due to a pause to determine pointing solutions. The flags below catch times just before and after when the amplitudes and phase are off. This flagging could also have been accomplished interactively in plotms.
# In CASA
default('flagdata')
flagdata(vis="g19_d2usb.ms",field="9",timerange='12:16:58~12:19:00')
flagdata(vis="g19_d2usb.ms",field="9",timerange='12:08:00~12:09:30')
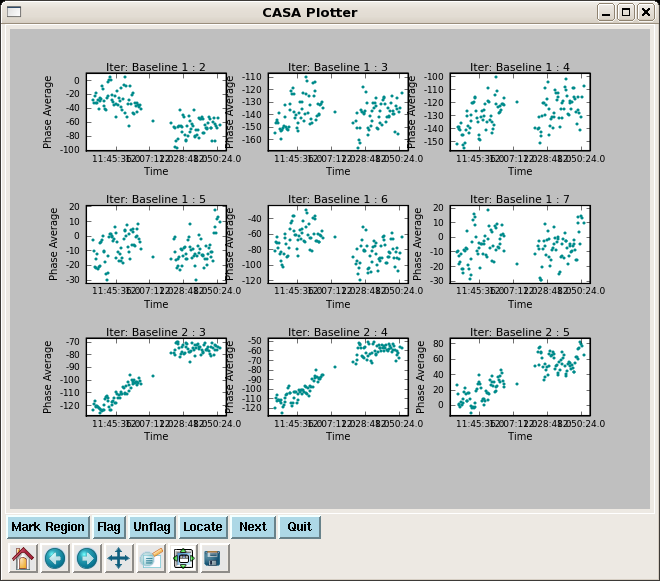
Here we plot phase vs. time for the bandpass calibrator 3c454.3 using plotxy:
# In CASA
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="phase",datacolumn="data",
iteration="baseline", antenna="",spw="0~23",field="9",
averagemode="vector",width="allspw",timebin="0",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
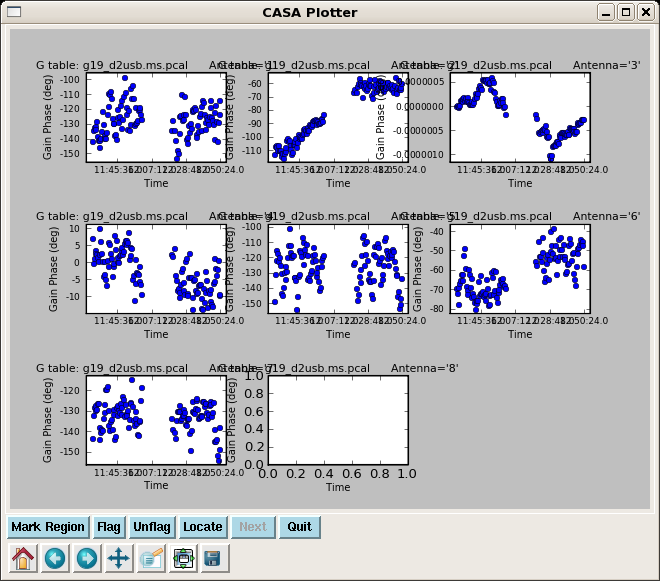
Start by determining an initial phase only solution for 3c454.3 using gaincal to take out major phase variations with time. For maximum sensitivity, we average all the spw together, with the exception of spw 4-7 which show large phase offsets from the other spw in phase vs. channel.
# In CASA
gaincal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.pcal',
field='9', spw='0~3,8~23', gaintype='G', minsnr=2.0,
refant='3', calmode='p',solint='int', combine='spw')
clearstat()
plotcal(caltable="g19_d2usb.ms.pcal",xaxis="time",yaxis="phase",
field="9",antenna="",spw="",timerange="",subplot=331,
overplot=False,clearpanel="Auto",iteration="antennas",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
Next we do a bandpass calibration. There are several possible solution types. A regular 'B' type solution can work well if you have lots of S/N on your bandpass calibrator. You may wish to make this table and compare to 'BPOLY' solution further down.
# In CASA
bandpass(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.bpcal',
field='9', spw='0~23', bandtype='B', solint='inf', combine='scan',
refant='3', gaincurve=False, opacity=0.0,
gaintable='g19_d2usb.ms.pcal',spwmap=[0])
plotcal(caltable="g19_d2usb.ms.bpcal",xaxis="freq",yaxis="amp",
field="9",antenna="",spw="",timerange="",subplot=311,
overplot=False,clearpanel="Auto",iteration="antenna",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
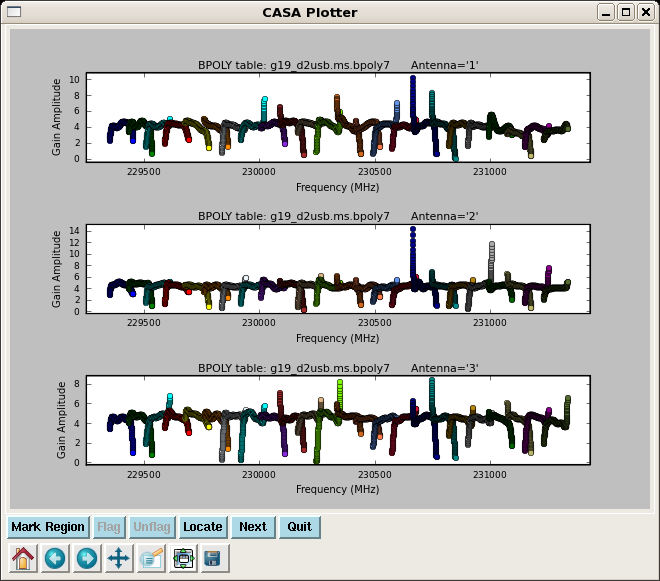
A polynomial bandpass solution is more sensitive than simply fitting a polynomial to the channel based solutions 'B' after the fact. For the mm/submm, where the S/N is often low on the bandpass calibrator, 'BPOLY' is often the best choice, but every dataset is different. You may especially need to play with the degamp and degphase parameters in bandpass. The parameter combine='scan' is needed to average together the two 3c454.3 scans. The spwmap=[0] is telling it to apply the phase solution that was found from the average of all spw to each spw before determining the bandpass solution.
# In CASA
bandpass(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.bpoly7',
field='9', spw='0~23', bandtype='BPOLY',
solint='inf', combine='scan',
refant='3', degamp=7, degphase=7,
gaintable='g19_d2usb.ms.pcal',spwmap=[0])
clearstat()
plotcal(caltable="g19_d2usb.ms.bpoly7",xaxis="freq",yaxis="amp",
field="9",antenna="",spw="",timerange="",subplot=311,
overplot=False,clearpanel="Auto",iteration="antenna",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
Unfortunately, this kind of plot with many spw and colors takes a long time. Be patient for now, as it's being worked on. You can safely ignore where the solution gets large at edges of spw. This is showing where we flagged channels - this is also being worked on. Zoom in if necessary to see that inner portions are nice and flat.
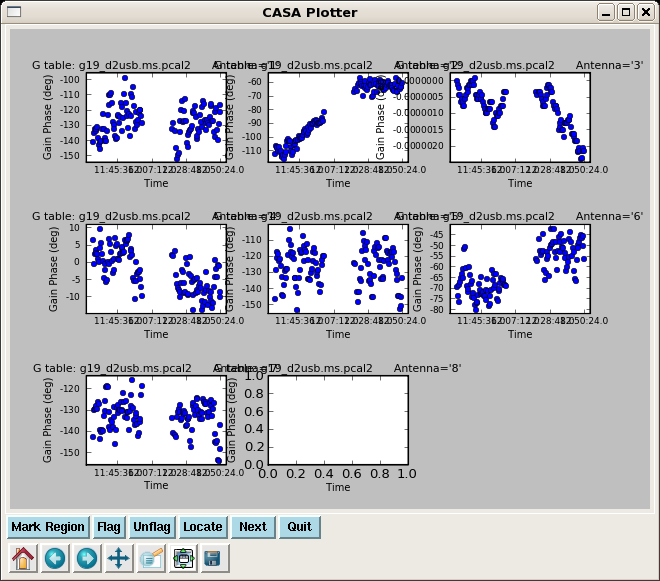
The next step is to apply the bandpass solution while re-calculating with gaincal the phase only solution for 3c454.3 on a short time interval (here, we use the integration time), but now including all spw (including 4~7 since antenna based solutions will have taken out phase vs.spw differences).
# In CASA
gaincal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.pcal2',
field='9', spw='0~23', gaintype='G', minsnr=2.0,
refant='3', calmode='p',solint='int', combine='spw',
gaintable=['g19_d2usb.ms.bpoly7'])
clearstat()
plotcal(caltable="g19_d2usb.ms.pcal2",xaxis="time",yaxis="phase",
field="9",antenna="",spw="",timerange="",subplot=331,
overplot=False,clearpanel="Auto",iteration="antennas",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
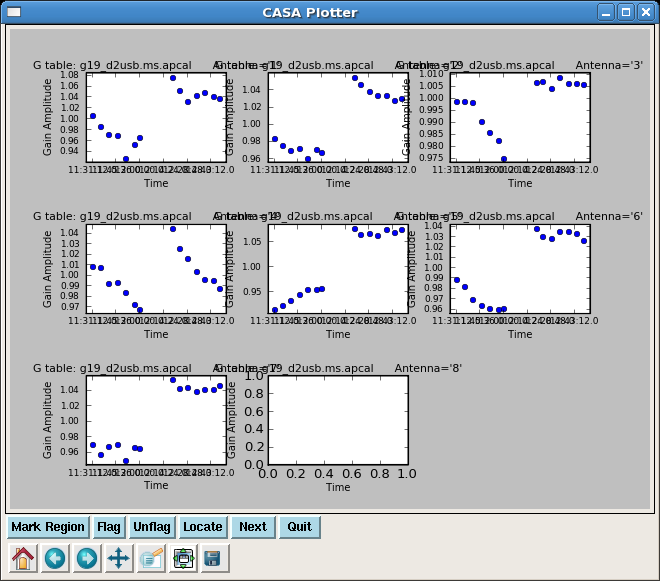
Next we apply the antenna based bandpass, and phase only solution to solve for amplitude solutions over a longer solint to increase the S/N. The spwmap=[[0],[]] tells gaincal that the pcal solution is independent of spw while the bpoly7 solution is per spw. spwmap order must be in the same order as the tables in gaintable.
# In CASA
gaincal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.apcal',
field='9', spw='0~23', gaintype='G', minsnr=2.0,
refant='3', calmode='a',solint='300s', combine='spw',
gaintable=['g19_d2usb.ms.pcal2','g19_d2usb.ms.bpoly7'],
spwmap=[[0],[]])
clearstat()
plotcal(caltable="g19_d2usb.ms.apcal",xaxis="time",yaxis="amp",
field="9",antenna="",spw="",timerange="",subplot=331,
overplot=False,clearpanel="Auto",iteration="antennas",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
Although in previous steps solutions were applied "on-the-fly", the data were not actually changed. Running applycal will fill the corrected data column in the ms. Again be careful with specification of gaintable, and keeping the same order in spwmap and gainfield. This applycal is not necessary for later steps, but allows plotting of data calibration to this point.
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='9',
gaintable=['g19_d2usb.ms.pcal2','g19_d2usb.ms.apcal',
'g19_d2usb.ms.bpoly7'],
spwmap=[[0],[0],[]],gainfield=['9','9','9'])
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="freq",yaxis="amp",
ydatacolumn="corrected",iteraxis="baseline",
spw="0~23",field="9",averagedata=True,avgtime='99999')
We can also view the data in a single plot using the slower plotxy:
# In CASA
plotxy(vis="g19_d2usb.ms",xaxis="channel",yaxis="amp",
datacolumn="corrected",iteration="baseline",
antenna='',spw="0~23",field="9",
averagemode="vector",width="1",timebin="all",crossscans=T,
crossbls=False,stackspw=False,extendflag=F,
extendchan="",extendspw="",extendant="",extendtime="",
plotcolor='darkcyan',subplot=331,
multicolor="none",figfile="")
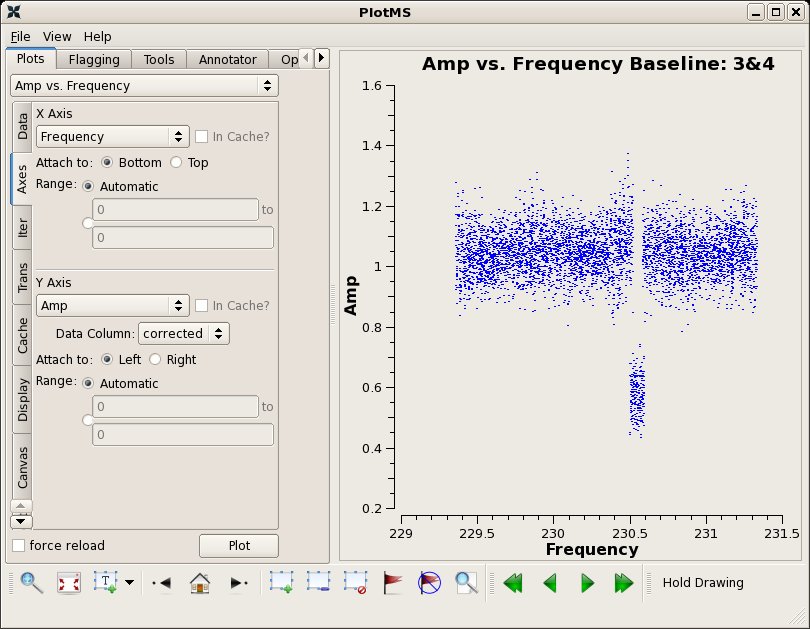
We can see by paging through the plots iterated over baseline (press the arrow button on the plotms display) that SMA data often show residual baseline based problems as a function of spw after antenna based bandpass (i.e. a few abnormally low amplitudes for a few spw on a few baselines). To fix this problem we need to do a baseline based amplitude only solution per spw, but we'll need to remove an overall normalization factor first by combining all spw (this option will be incorporated into blcal one day so that only one run is necessary). Note the significant change on baseline 1-6 in the following call to plotcal.
# In CASA
blcal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.blcal1',
field='9', spw='0~23',solint='inf', combine='spw,scan',
gaintable=['g19_d2usb.ms.pcal2','g19_d2usb.ms.apcal',
'g19_d2usb.ms.bpoly7'],
calmode='a',spwmap=[[0],[0],[]])
blcal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.blcal2',
field='9', spw='0~23',solint='inf', combine='scan',
gaintable=['g19_d2usb.ms.pcal2','g19_d2usb.ms.apcal',
'g19_d2usb.ms.blcal1','g19_d2usb.ms.bpoly7',],
calmode='a',spwmap=[[0],[0],[0],[]])
clearstat()
plotcal(caltable="g19_d2usb.ms.blcal2",xaxis="freq",yaxis="amp",
field="9",antenna="",spw="",timerange="",subplot=511,
overplot=False,clearpanel="Auto",iteration="antenna",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
Now we can do a new applycal including only the second blcal2 solution. Note that applycal overwrites the corrected column, so there is no need to clearcal. Like the antenna based bandpass the blcal solution is independent for each spw, so we set spwmap = [].
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='9',
gaintable=['g19_d2usb.ms.pcal2','g19_d2usb.ms.apcal',
'g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],
spwmap=[[0],[0],[],[]],gainfield=['9','9','9','9'])
Note how flat the full bandpass is after all this calibration (be sure to view all baseline iterations using the plotms arrow button):
# In CASA
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="freq",yaxis="amp",
ydatacolumn="corrected",iteraxis="baseline",
spw="0~23",field="9",averagedata=True,avgtime='99999')
Inspection of gain calibrators and setting the flux scale
Apply antenna and baseline based bandpass solutions to the gain calibrators with applycal:
# In CASA
clearstat()
applycal(vis='g19_d2usb.ms',spw='0~23', field='1,6',
gaintable=['g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],
spwmap=[[],[]],gainfield=['9','9'])
Have a look at the corrected data for both calibrators with channel vs. amplitude:
# In CASA
# First field 1:
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="channel",yaxis="amp",datacolumn="corrected",
iteration="baseline", antenna="",spw="",field="1",
averagemode="vector",width="1",timebin="all",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
# Next field 6:
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="channel",yaxis="amp",datacolumn="corrected",
iteration="baseline", antenna="",spw="",field="6",
averagemode="vector",width="1",timebin="all",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
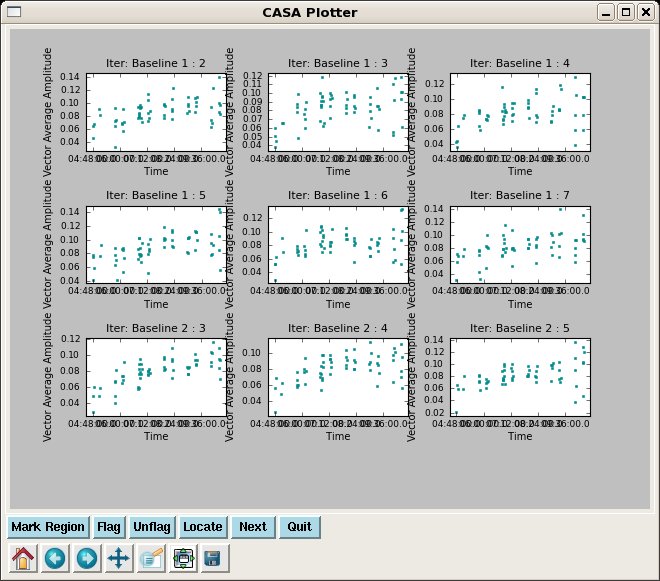
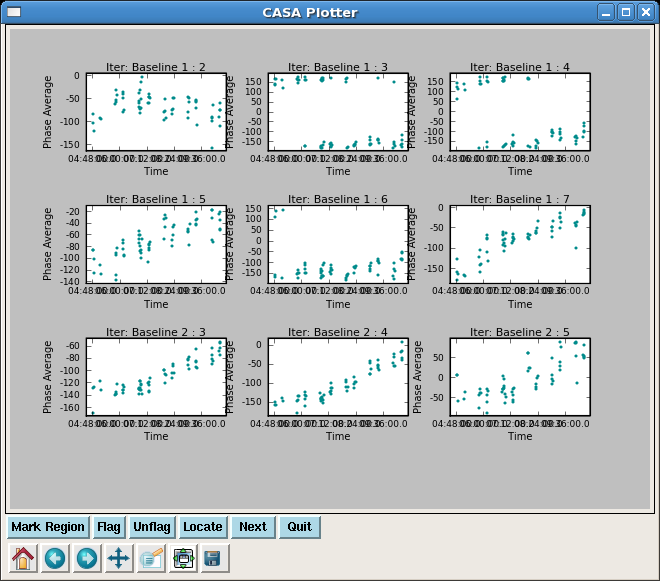
Have a look at the corrected data for both calibrators in phase and amplitude vs. time. We can't color by field in plotxy, unfortunately, so this is easiest to do separately.
# In CASA
# First, amplitude vs. time for field 1:
plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",datacolumn="corrected",
iteration="baseline", antenna="",spw="0~23",field="1",
averagemode="vector",width="allspw",timebin="0",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
# Second, amplitude vs. time for field 6:
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",datacolumn="corrected",
iteration="baseline", antenna="",spw="0~23",field="6",
averagemode="vector",width="allspw",timebin="0",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="red",
overplot=F,showflags=False,interactive=True,figfile="")
# Third, phase vs. time for field 1:
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="phase",datacolumn="corrected",
iteration="baseline", antenna="",spw="0~23",field="1",
averagemode="vector",width="allspw",timebin="0",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
# Last, phase vs. time for field 6:
clearstat()
plotxy(vis="g19_d2usb.ms",xaxis="time",yaxis="phase",datacolumn="corrected",
iteration="baseline", antenna="",spw="0~23",field="6",
averagemode="vector",width="allspw",timebin="0",
subplot=331,plotsymbol=".",
multicolor="none",plotcolor="darkcyan",
overplot=False,showflags=False,interactive=True,figfile="")
At the time of these observations, the calibrator J1911 had the most recent and relatively stable monitor data (SMA website), so we will use it for absolute flux calibration. In future tools for using resolved planets will be implemented.
1911-201 1mm 17 Jul 2005 11:38 SMA 274.5 1.88 +/- 0.13 mgurwell 1mm 05 Sep 2005 08:10 SMA 221.4 1.99 +/- 0.12 mgurwell
# In CASA
setjy(vis='g19_d2usb.ms', field='6', spw='0~23', fluxdensity=[2.0,0.,0.,0.])
Final gain and flux calibration for all calibrators
Next, we calculate two separate phase solutions from the data. To avoid decorrelation of the amplitude, the phase solutions fed to the amplitude calibration step should be at the shortest solint your S/N can support. Typically this will be the integration time.
# In CASA
gaincal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.allpcal',
field='1,6,9', spw='0~23', gaintype='G', minsnr=2.0,
refant='3', calmode='p',solint='int', combine='spw',
gaintable=['g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],
spwmap=[[],[]])
Unless you chose to do some more sophisticated smoothing, the phase solutions applied to your target might as well be on the scan time of your phase calibrator(s). Without combine='scan', the solution will not cross scan boundaries even though solint='inf'. This is an easy way to "manually" interpolate.
# In CASA
gaincal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.allpcalscan',
field='1,6,9', spw='0~23', gaintype='G', minsnr=2.0,
refant='3', calmode='p',solint='inf', combine='spw',
gaintable=['g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],
spwmap=[[],[]])
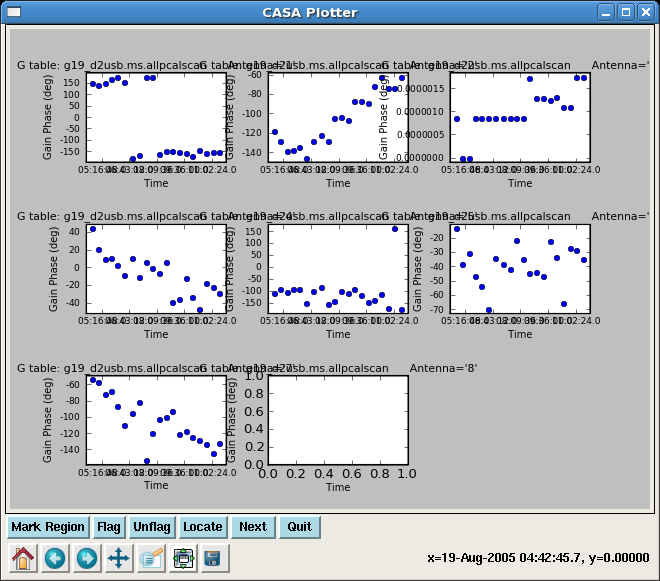
Plot the phase solutions of the phase calibrators. Can repeat for both short and long solution intervals.
# In CASA
clearstat()
plotcal(caltable="g19_d2usb.ms.allpcalscan",xaxis="time",yaxis="phase",
field="1,6",antenna="",spw="",timerange="",subplot=331,
overplot=False,clearpanel="Auto",iteration="antennas",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
Amplitude calibration with longer solint and applying short solint phase solution.
# In CASA
gaincal(vis='g19_d2usb.ms', caltable='g19_d2usb.ms.allapcal',
field='1,6,9', spw='0~23', gaintype='G', minsnr=2.0,
refant='3', calmode='ap',solint='300s', combine='spw',
gaintable=['g19_d2usb.ms.allpcal','g19_d2usb.ms.bpoly7',
'g19_d2usb.ms.blcal2'],spwmap=[[0],[],[]])
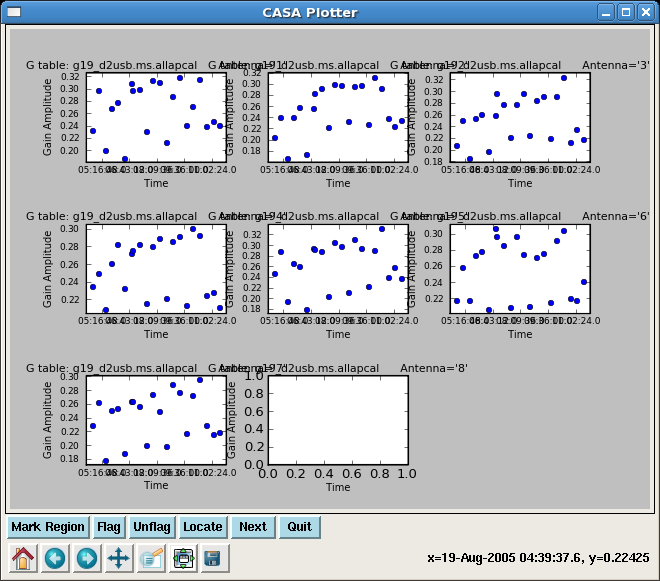
Plot the amplitude solutions:
# In CASA
clearstat()
plotcal(caltable="g19_d2usb.ms.allapcal",xaxis="time",yaxis="amp",
field="1,6",antenna="",spw="",timerange="",subplot=331,
overplot=False,clearpanel="Auto",iteration="antennas",
plotsymbol="o",plotcolor="blue",showgui=True,figfile="")
Derive the absolute flux calibration using fluxscale (based on setjy above):
# In CASA
fluxscale(vis='g19_d2usb.ms',caltable='g19_d2usb.ms.allapcal',
fluxtable='g19_d2usb.ms.fluxcal',reference='6')
Apply final calibrations
The final application of calibration for each source is done independently below. Flux densities from SMA flux monitoring are provided as checks of the absolute flux calibration.
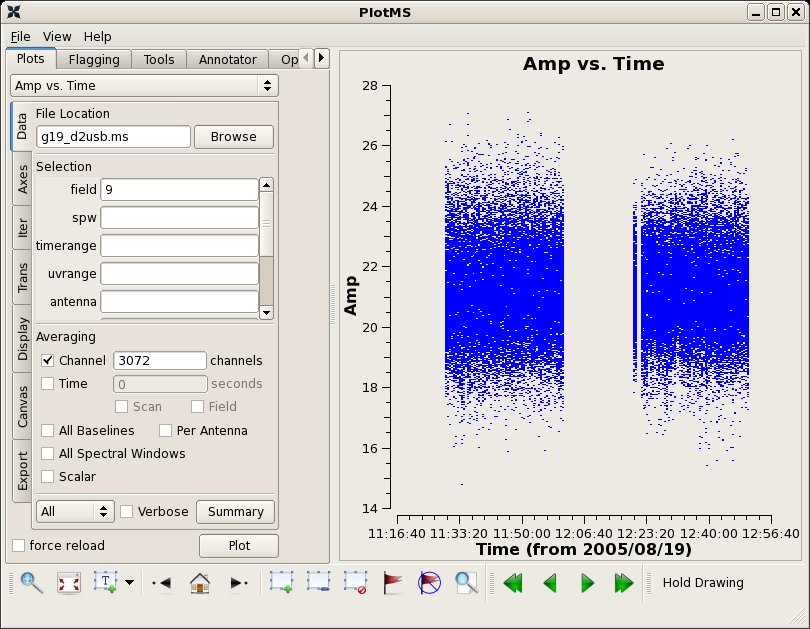
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='9',
gaintable=['g19_d2usb.ms.allpcal','g19_d2usb.ms.fluxcal',
'g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],spwmap=[[0],[0],[],[]],
gainfield=['9','9','9','9'])
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",field='9',
avgchannel='3072',ydatacolumn='corrected')
3C454.3 1mm 19 Aug 2005 16:01 SMA 225.3 27.36 +/- 1.37 mgurwell
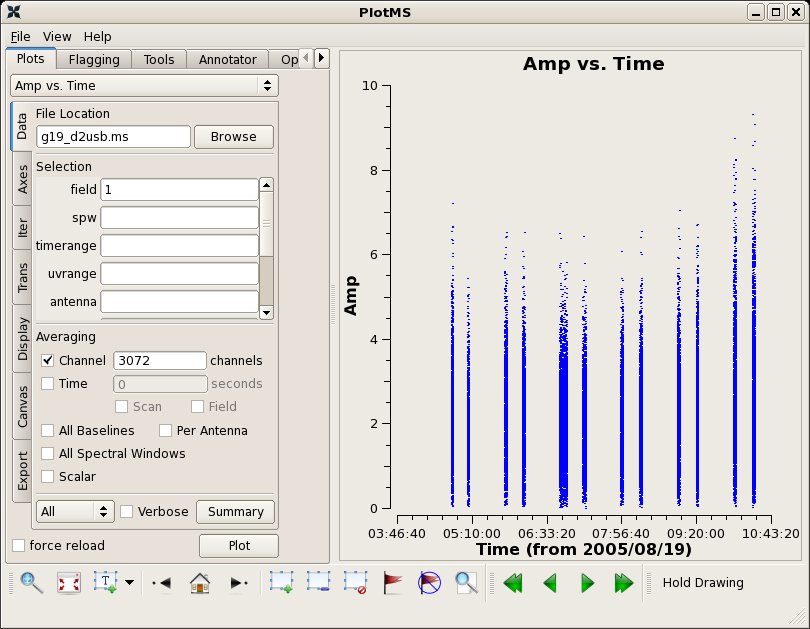
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='1',
gaintable=['g19_d2usb.ms.allpcal','g19_d2usb.ms.fluxcal',
'g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],spwmap=[[0],[0],[],[]],
gainfield=['1','1','9','9'])
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",field='1',
avgchannel='3072',ydatacolumn='corrected')
J1743-038 1mm 26 Aug 2005 06:05 SMA 225.6 1.71 +/- 0.09 mgurwell
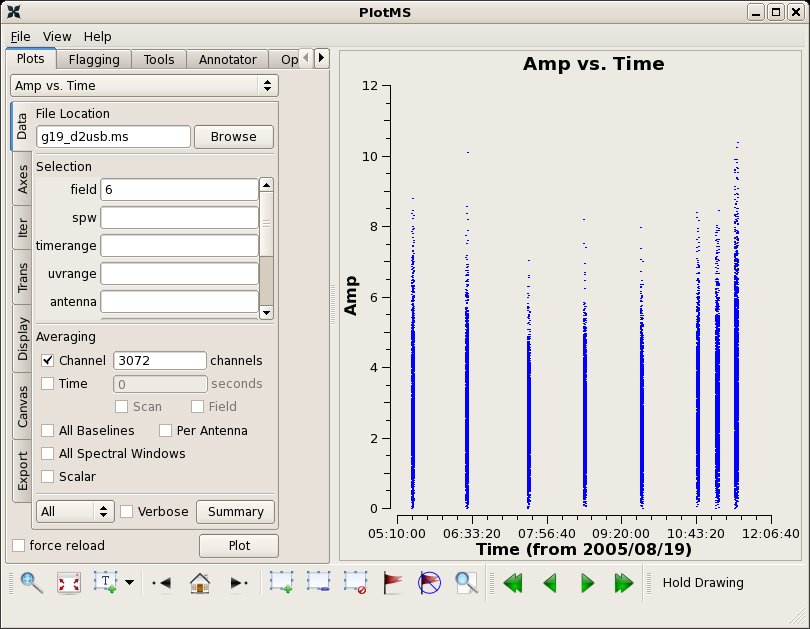
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='6',
gaintable=['g19_d2usb.ms.allpcal','g19_d2usb.ms.fluxcal',
'g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],spwmap=[[0],[0],[],[]],
gainfield=['6','6','9','9'])
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="time",yaxis="amp",field='6',
avgchannel='3072',ydatacolumn='corrected')
J1911-201 1mm 17 Jul 2005 11:38 SMA 274.5 1.88 +/- 0.13 mgurwell 1mm 05 Sep 2005 08:10 SMA 221.4 1.99 +/- 0.12 mgurwell
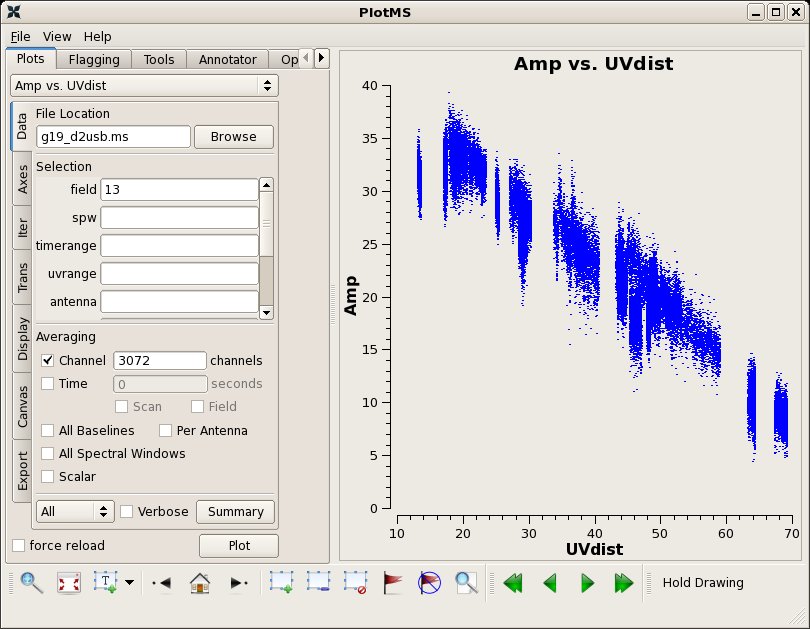
Since Uranus is resolved we apply the solutions from 3C454.3 to achieve at least some reduction in instrumental gain variations.
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='13',
gaintable=['g19_d2usb.ms.allpcal','g19_d2usb.ms.fluxcal',
'g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],spwmap=[[0],[0],[],[]],
gainfield=['9','9','9','9'])
clearstat()
plotms(vis="g19_d2usb.ms",xaxis="uvdist",yaxis="amp",field='13',
avgchannel='3072',ydatacolumn='corrected')
A UV-distance plot of the expected flux of Uranus can be obtained from the SMA webpage. In future we will implement tools to use planets for absolute flux calibration.
# In CASA
applycal(vis='g19_d2usb.ms',spw='0~23', field='2,3,4,5,7,8',
gaintable=['g19_d2usb.ms.allpcalscan','g19_d2usb.ms.fluxcal',
'g19_d2usb.ms.bpoly7','g19_d2usb.ms.blcal2'],spwmap=[[0],[0],[],[]],
gainfield=['1,6','1,6','9','9'])
Split off target data for further inspection
# In CASA
split(vis="g19_d2usb.ms",outputvis='g19_d2usb_targets.ms',
field="2,3,4,5,7,8")
To reinitialize the scratch columns for use by later tasks, we need to run clearcal for the new dataset. Then run listobs to check the target data names.
# In CASA
clearcal(vis='g19_d2usb_targets.ms')
listobs(vis="g19_d2usb_targets.ms",verbose=F)
ID Code Name Right Ascension Declination Epoch 0 g19.3a 18:25:58.70 -12.03.57.80 J2000 1 g19.3b 18:25:57.40 -12.04.18.50 J2000 2 g19.3c 18:25:56.09 -12.04.28.20 J2000 3 g19.3d 18:25:54.60 -12.04.23.10 J2000 4 g19.3e 18:25:54.80 -12.04.43.80 J2000 5 g19.3f 18:25:53.30 -12.04.51.00 J2000
Tip: After splitting, the "corrected" data column in the original vis file will become the "data" and "corrected" columns and the field IDs are renumbered.
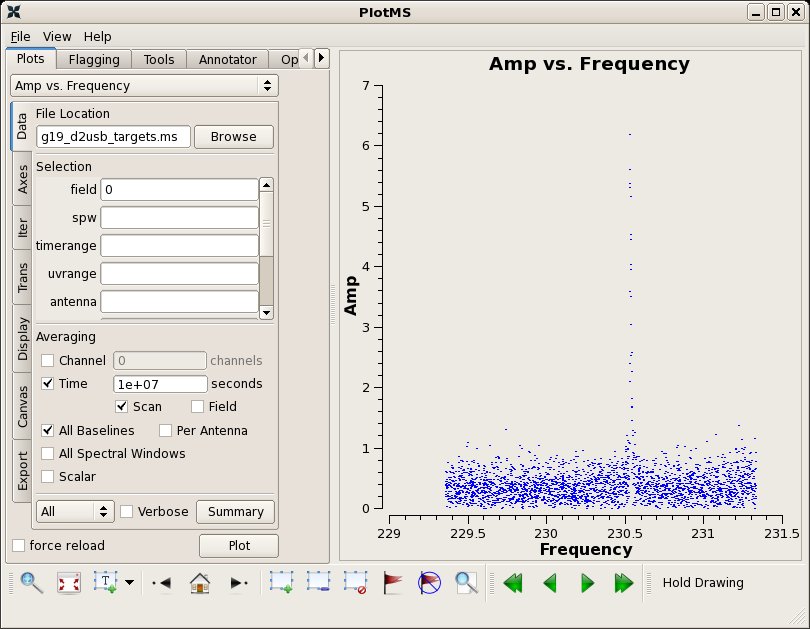
# In CASA
clearstat()
plotms(vis='g19_d2usb_targets.ms',xaxis="frequency",yaxis="amp",field='0',
avgtime='1e7',avgscan=True,avgbaseline=True)
clearstat()
plotms(vis='g19_d2usb_targets.ms',xaxis="time",yaxis="amp",field='',
avgchannel='3072',avgspw=True)
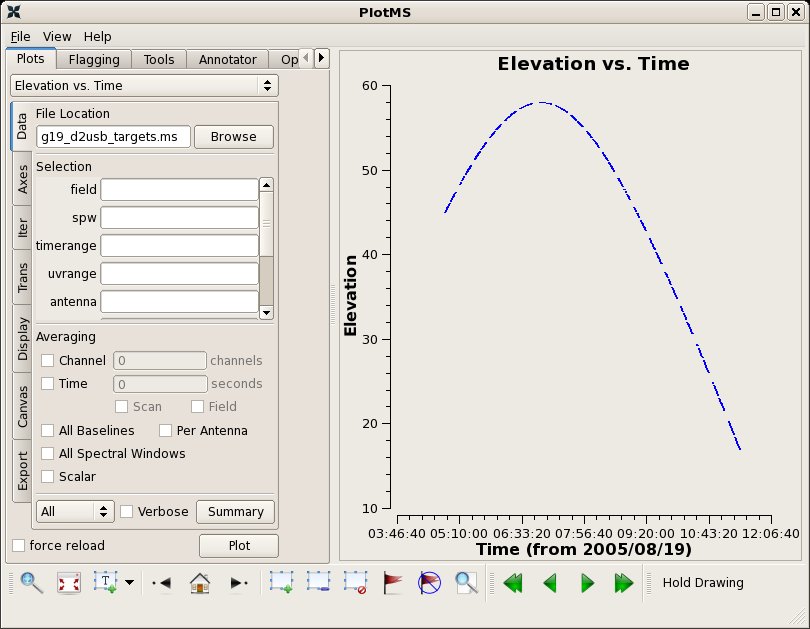
Data at times > 11:04:00 show lots of scatter: the elevation of these data are very low and are worth flagging. One can plotms with xaxis="time", yaxis="elevation" to see this.
# In CASA
clearstat()
plotms(vis='g19_d2usb_targets.ms',xaxis="time",yaxis="elevation",field='')
# In CASA
flagdata(vis="g19_d2usb_targets.ms",field="",timerange='>11:04:00')
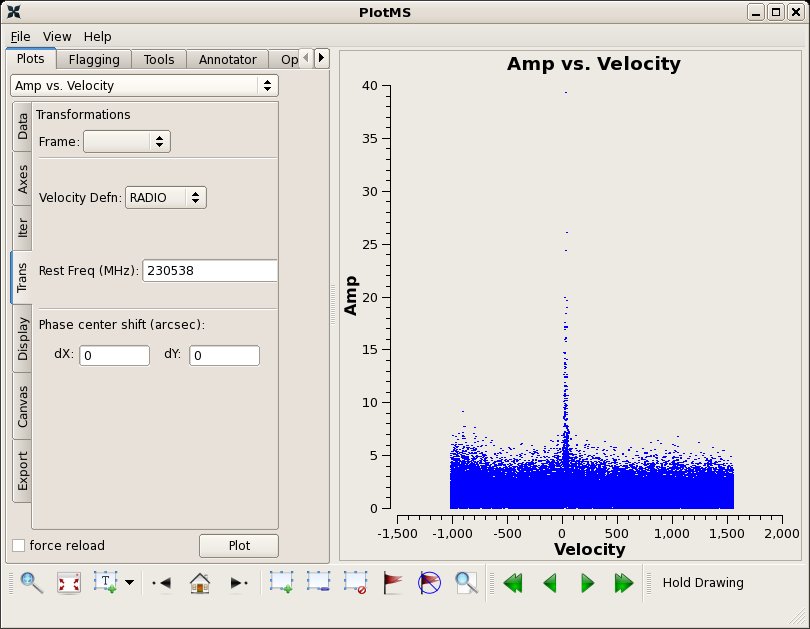
Note that because the Doppler tracked frequency was 13CO (+ 10GHz in the USB), the velocity diplayed in plotms will not be correct unless you set the rest frequency to the line of interest. You can do this in the plotms call, or in the Trans tab in the plotms display:
CO(2-1) restfreq=230.53797 GHz
# In CASA
clearstat()
plotms(vis="g19_d2usb_targets.ms",xaxis="velocity",yaxis="amp",
ydatacolumn="data",selectdata=True,
spw="0~23",field="0",
averagedata=True,avgtime='99999',avgscan=True,
transform=True,restfreq="230.53797GHz")
The other weak line detected in this sideband is not visible in a vector average plot.
A-CH3OH (8 -1 8 0 -- 7 +0 7 0) 229.75876 GHz
Continuum imaging and subtraction
Next we want to make a continuum-free line dataset. We use field 0 to pick the line free spw, and can see that the line is confined to spws 13~15 by colorizing by spw:
# In CASA
clearstat()
plotms(vis="g19_d2usb_targets.ms",xaxis="freq",yaxis="amp",
ydatacolumn="data",selectdata=True,coloraxis="spw",
spw="0~23",field="0",averagedata=True,avgtime='99999')
Copy the original ms in case something goes wrong. Currently uvcontsub2 will change the input model and corrected columns, so it's best to make a back up. This behavior will change in future.
# In CASA
os.system('cp -r g19_d2usb_targets.ms g19_d2usb_targets_line.ms')
Now use new task uvcontsub2 to subtract the continuum. Unlike uvcontsub, this task will subtract the continuum estimate for spw that are not in the fitspw list. In future releases uvcontsub2 will likely replace uvcontsub. Setting fitspw='0~12,16~23' tells uvcontsub2 where to fit the continuum emission.
# In CASA
uvcontsub2(vis='g19_d2usb_targets_line.ms',fitspw='0~12,16~23',
solint='int',combine='spw',spw='0~23',
fitorder=1)
The directory g19_d2usb_targets_line.ms.contsub/ contains the continuum-subtracted line data. We will make a separate dataset, g19_d2usb_targets_line.ms.cont, using the split task for imaging. Because the continuum model subtracted from the line dataset by uvcontsub2 is necessarily a smoothed fit, images made with it are liable to have their field of view reduced in some strange way. Images of the continuum should be made by simply excluding the line channels, as we show here.
# In CASA
split(vis='g19_d2usb_targets.ms',outputvis='g19_d2usb_targets_line.ms.cont',
datacolumn='data',field='',spw='0~12,16~23')
Next we image the continuum data:
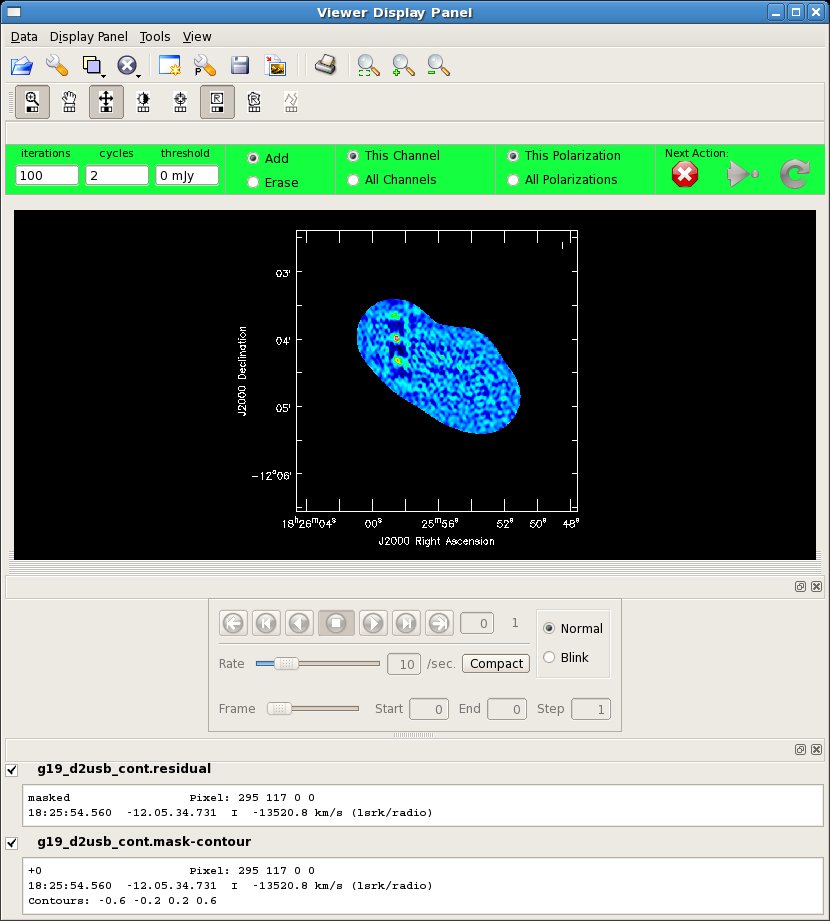
Tip: for sparse mosaics like this one, the subparameter ftmachine='ft' will typically give better results than the default ftmachine='mosaic'. For Nyquist sampled mosaics, ftmachine='mosaic' is the best choice, and the only choice for heterogeneous arrays. Also note that in a mosaic, the rms noise changes as a function of position. Therefore, the rms noise that you calculate from the radiometer equation is only valid where the combined primary beam response of the mosaic is near one. clean weights the decision of what to clean next by the primary beam response function (PBRF), so emission near PBRF=1 will be cleaned faster than elsewhere. Thus, if you have emission away from the "sweet spot(s)" you will need to clean to a threshold lower than you might expect from the anticipated rms noise. Development is on-going to provide other options which may work better for the case of strong emission near image boundaries.
# In CASA
clean(vis='g19_d2usb_targets_line.ms.cont',imagename='g19_d2usb_cont',
field='',spw='0~23',
mode='mfs',niter=200,gain=0.1,threshold=0.0,
psfmode='clark',imagermode='mosaic',scaletype='SAULT',
ftmachine='ft',
interactive=T,imsize=500,cell="0.5arcsec",
phasecenter="J2000 18h25m56.09 -12d04m28.20",
pbcor=F,minpb=0.2)
viewer('g19_d2usb_cont.image')
Here you can see one stronger continuum source to the NE (18:25:58.5 -12:03:58.9) and one weak one to the SW (18:25:52, -12:04:53). The weak one is only visible after one round of cleaning. Because the mosaic is sparse you should make a clean mask to get the best cleaned image.
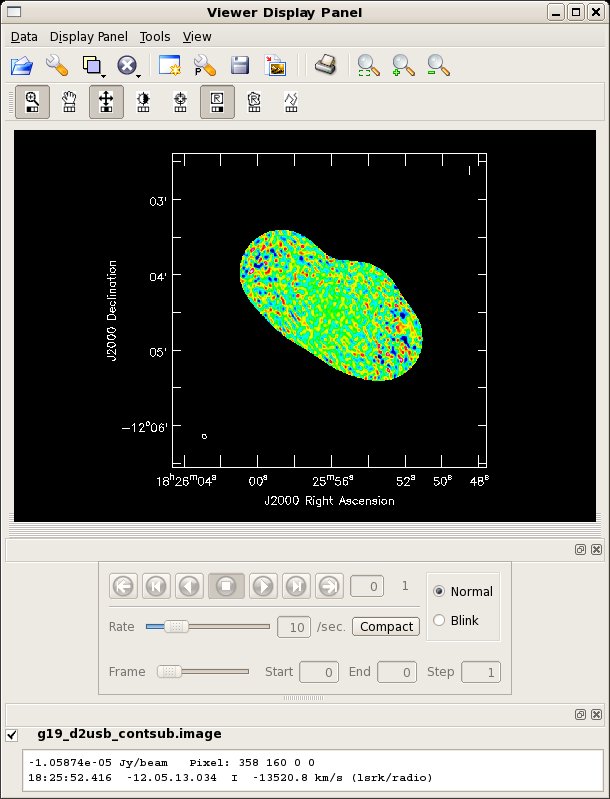
Make a dirty image of the continuum subtracted data, excluding the spws which contain line emission. You should see nothing but noise.
# In CASA
clean(vis='g19_d2usb_targets_line.ms.contsub',imagename='g19_d2usb_contsub',
field='',spw='0~12,16~23',
mode='mfs',niter=0,gain=0.1,threshold=0.0,
psfmode='clark',imagermode='mosaic',scaletype='SAULT',
ftmachine='ft',
interactive=F,imsize=500,cell="0.5arcsec",
phasecenter="J2000 18h25m56.09 -12d04m28.20",
pbcor=F,minpb=0.2)
You can look at the image with the viewer; you should see nothing because the continuum has been subtracted.
# In CASA
viewer('g19_d2usb_contsub.image')
Spectral line imaging
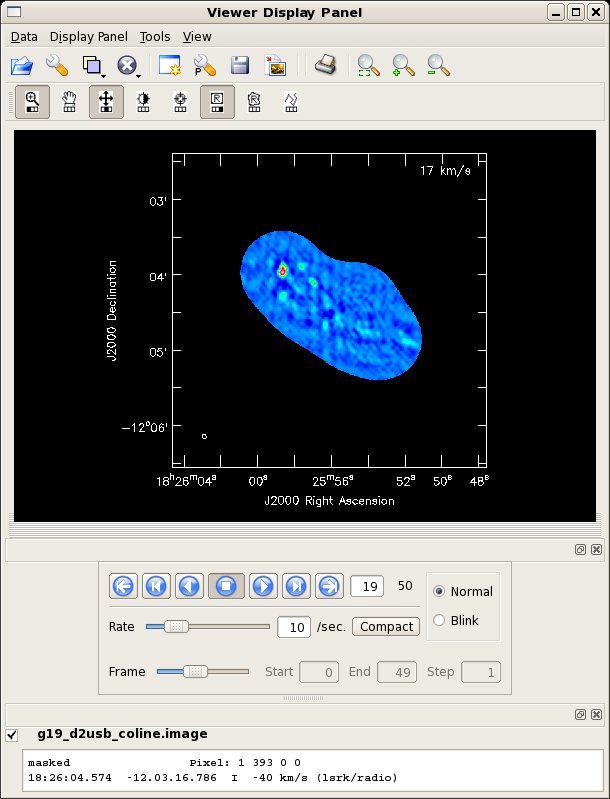
This image selects a channel range suitable to image the 12CO(2-1) line. Notice the use of the restfreq parameter to get the velocity scale correct.
# In CASA
clean(vis='g19_d2usb_targets_line.ms.contsub',imagename='g19_d2usb_coline',
field='',spw='',
mode='velocity',start='-40km/s',nchan=50,width='3km/s',
interpolation='linear',
niter=1000,gain=0.1,threshold=0.2,
psfmode='clark',imagermode='mosaic',scaletype='SAULT',
ftmachine='ft',restfreq='230.53797GHz',
interactive=T,npercycle=400,
imsize=500,cell="0.5arcsec",
phasecenter="J2000 18h25m56.09 -12d04m28.20",
pbcor=F,minpb=0.15)
It is essential to run with interactive=T and make a clean mask. A clean mask is available in the original data directory spw_fits_files. To use the pre-made mask, run:
# In CASA
clean(vis='g19_d2usb_targets_line.ms.contsub',imagename='g19_d2usb_coline',
field='',spw='',
mode='velocity',start='-40km/s',nchan=50,width='3km/s',
niter=1000,gain=0.1,threshold=0.2,
psfmode='clark',imagermode='mosaic',scaletype='SAULT',
ftmachine='ft',restfreq='230.53797GHz',
interactive=F,npercycle=400,
imsize=500,cell="0.5arcsec",
mask='spw_fits_files/coline.mask',
phasecenter="J2000 18h25m56.09 -12d04m28.20",
pbcor=F,minpb=0.15)
View the image produced:
# In CASA
viewer('g19_d2usb_coline.image')
Now make 0th and 1st moment maps.
# In CASA
# 0th moment map
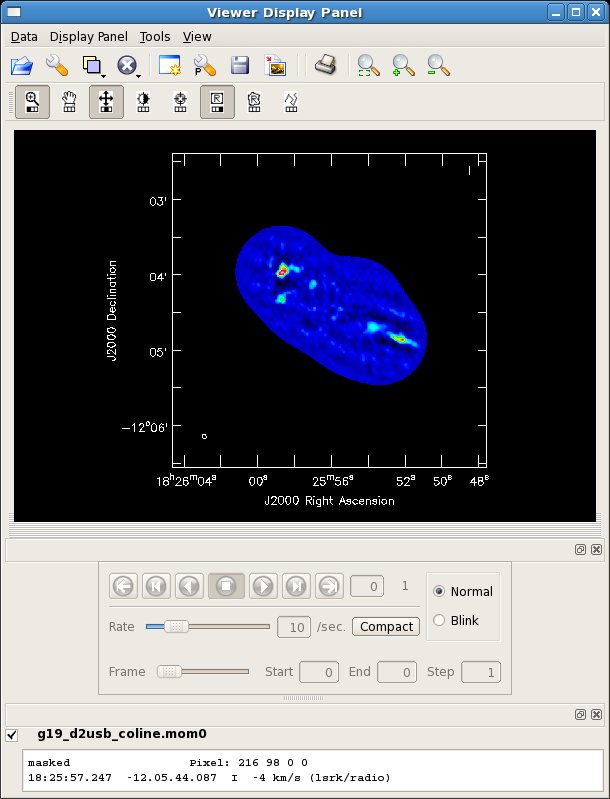
immoments(imagename='g19_d2usb_coline.image',moments=[0],axis='spectral',
chans='12~39',outfile='g19_d2usb_coline.mom0')
viewer('g19_d2usb_coline.mom0')
# 1st moment map
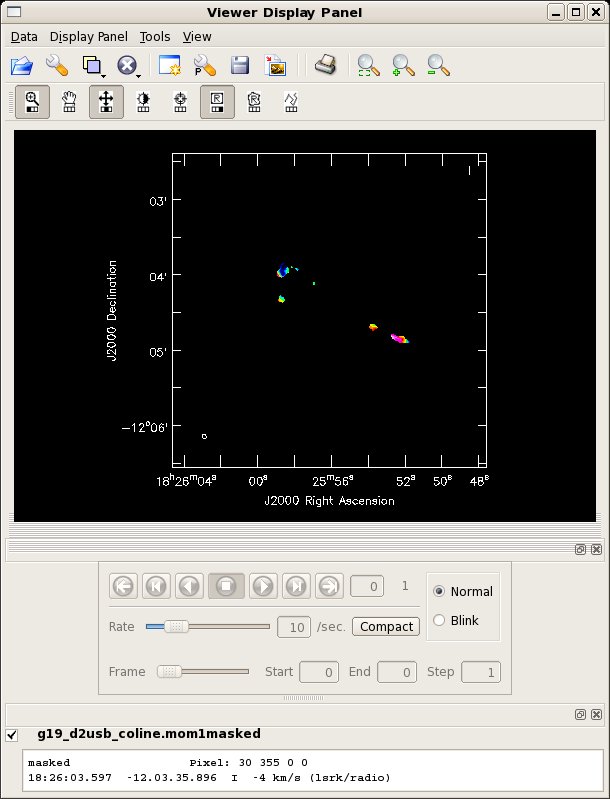
immoments(imagename='g19_d2usb_coline.image',moments=[1],axis=3,
chans='12~39',outfile='g19_d2usb_coline.mom1',
includepix=[0,2])
immath(outfile='g19_d2usb_coline.mom1masked',mode='evalexpr',
expr='"g19_d2usb_coline.mom1"["g19_d2usb_coline.mom0">9]')
viewer('g19_d2usb_coline.mom1masked')
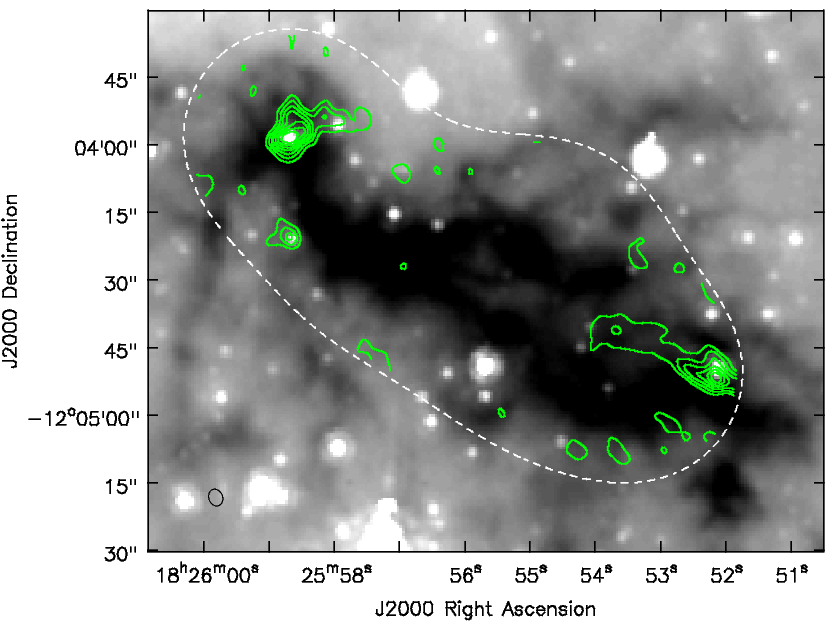
By clicking on the "folder" icon in the viewer window you can bring up the moment0 as a contour image. The strange appearance of the the moment1 map toward the NE source is probably due to more than one outflow being superposed.
Note that one could and should correct the image for the primary beam response using immath to divide the image by the .flux image. One would then need to make a mask for the moment images in order to hide the ugly stuff at the edges.
Last checked on CASA Version 3.2.0.
--Rfriesen 10:25, 5 May 2011 (PDT)