IRAS16293 Band9 - Imaging for CASA 6.6.1: Difference between revisions
| (25 intermediate revisions by the same user not shown) | |||
| Line 227: | Line 227: | ||
=== Selfcal Round 2 === | === Selfcal Round 2 === | ||
Now you are ready for the second cleaning. For this one we are using the mask resulting from the previous cleaning. In the next blocks of commands we will repeat this process (clean, | Now you are ready for the second cleaning. For this one we are using the mask resulting from the previous cleaning. In the next blocks of commands we will repeat this process (clean, carta, gaincal, plotms, applycal, clean, etc.) to improve the overall quality of the image. You can add to your clean mask as you go, but be careful to avoid adding sidelobe emission as real structure. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 283: | Line 283: | ||
# In CASA | # In CASA | ||
# Cleaning 3 | # Cleaning 3 | ||
os.system('rm -rf | os.system('rm -rf IRAS16293_Band9.calibrated.cont_p2*') | ||
tclean(vis='IRAS16293_Band9. | tclean(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms', | ||
imagename='IRAS16293_Band9. | imagename='IRAS16293_Band9.calibrated.cont_p2', | ||
mask='IRAS16293_Band9. | mask='IRAS16293_Band9.calibrated.cont_p1.mask', | ||
spw='0~3',field='',phasecenter=3, | spw='0~3',field='',phasecenter=3, | ||
specmode='mfs',gridder='mosaic',mosweight=True, | specmode='mfs',gridder='mosaic',mosweight=True, | ||
| Line 295: | Line 295: | ||
</source> | </source> | ||
Open "IRAS16293_Band9. | Open "IRAS16293_Band9.calibrated.cont_p2.image" in CARTA. There is very little change; the bold could try another iteration of phase self-cal using a shorter solint. For now, let's move on to amplitude. | ||
[[File:cont_apcal.png|200px|thumb|right|'''Fig. 6.''' Amplitude self-calibration solutions.]] | [[File:cont_apcal.png|200px|thumb|right|'''Fig. 6.''' Amplitude self-calibration solutions.]] | ||
| Line 305: | Line 305: | ||
# For amplitude self-cal we only want to get one solution per spw, per dataset | # For amplitude self-cal we only want to get one solution per spw, per dataset | ||
os.system('rm -rf cont_apcal') | os.system('rm -rf cont_apcal') | ||
gaincal(vis='IRAS16293_Band9. | gaincal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms', | ||
caltable='cont_apcal',gaintype='T',refant='DV14', | |||
calmode='ap',combine='scan,field',spw='',field='', | |||
solint='inf',minsnr=3.0,minblperant=4,gaintable='cont_pcal2') | |||
# ploting the table | # ploting the table | ||
plotms(vis='cont_apcal',xaxis='time',yaxis='amp', | plotms(vis='cont_apcal',xaxis='time',yaxis='amp', | ||
spw='',iteraxis='antenna',gridrows=5,gridcols=1, | |||
plotrange=[0,0,0.5,2],antenna='',timerange='', | |||
showgui=True) | |||
# and applying to the data | # and applying to the data | ||
applycal(vis='IRAS16293_Band9. | applycal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',spwmap=[], | ||
spw='',gaintable=['cont_pcal2','cont_apcal'],calwt=False, | |||
flagbackup=False) | |||
# A large correction is present for the 2nd dataset. This | # A large correction is present for the 2nd dataset. This | ||
# error was causing many of the artifacts noticeable in the previous | # amplitude error was causing many of the artifacts noticeable | ||
# in the previous images. | |||
</source> | </source> | ||
=== Final Aggregate Continuum Image === | === Final Aggregate Continuum Image === | ||
Continuing the final cleaning step. We will re-use the mask from the | Continuing the final cleaning step. We will re-use the mask from the cont_p2 image, and also set pbcor=True to output a pb-corrected image. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
# Cleaning 4 | # Cleaning 4 | ||
os.system('rm -rf | os.system('rm -rf IRAS16293_Band9.calibrated.cont_ap.*') | ||
tclean(vis='IRAS16293_Band9. | tclean(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms', | ||
imagename='IRAS16293_Band9. | imagename='IRAS16293_Band9.calibrated.cont_ap', | ||
spw='0~3',field='',phasecenter=3, | spw='0~3',field='',phasecenter=3, | ||
specmode='mfs',gridder='mosaic',mosweight=True, | specmode='mfs',gridder='mosaic',mosweight=True, | ||
imsize=720,cell='0.028arcsec', | imsize=720,cell='0.028arcsec', | ||
interactive=True,deconvolver='hogbom',nterms=1, | interactive=True,deconvolver='hogbom',nterms=1, | ||
mask='IRAS16293_Band9. | mask='IRAS16293_Band9.calibrated.cont_p2.mask', | ||
weighting='briggs',robust=0.5,savemodel='modelcolumn', | weighting='briggs',robust=0.5,savemodel='modelcolumn', | ||
niter=1000, threshold='40.0mJy',pbcor=True) | niter=1000, threshold='40.0mJy',pbcor=True) | ||
</source> | </source> | ||
Open "IRAS16293_Band9. | Open "IRAS16293_Band9.calibrated.cont_ap.image" in CARTA. | ||
<!-- HAS: I used 4 major cycles. --> | |||
We will use {{immath_6.6.1}} to cut out a sub-region of the cube because the area covered is large compared to the area of interest, and we want to keep the file size of the result as small as possible. | We will use {{immath_6.6.1}} to cut out a sub-region of the cube because the area covered is large compared to the area of interest, and we want to keep the file size of the result as small as possible. | ||
| Line 348: | Line 352: | ||
<source lang="python"> | <source lang="python"> | ||
# in CASA | # in CASA | ||
immath('IRAS16293_Band9. | immath('IRAS16293_Band9.calibrated.cont_ap.image.pbcor',outfile='IRAS16293_Band9.calibrated.cont_ap.image.subim.pbcor', | ||
box='221,160,493,430',expr='IM0') | box='221,160,493,430',expr='IM0') | ||
</source> | </source> | ||
| Line 363: | Line 367: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
exportfits(imagename='IRAS16293_Band9. | exportfits(imagename='IRAS16293_Band9.calibrated.cont_ap.image.subim.pbcor', | ||
fitsimage='IRAS16293_Band9. | fitsimage='IRAS16293_Band9.calibrated.cont_ap.image.subim.pbcor.fits') | ||
</source> | </source> | ||
| Line 372: | Line 376: | ||
# In CASA | # In CASA | ||
# Restore the flags made for the continuum imaging | # Restore the flags made for the continuum imaging | ||
flagmanager(vis='IRAS16293_Band9.calibrated.ms',mode='restore',versionname='Original') | flagmanager(vis='IRAS16293_Band9.calibrated.tavg.ms',mode='restore',versionname='Original') | ||
</source> | </source> | ||
Then we need to subtract the continuum, using the line free channels. The line free channels are just the inverse of the line channels identified earlier. | Then we need to subtract the continuum, using the line free channels. The line free channels are just the inverse of the line channels identified earlier. We will use {{uvcontsub_6.6.1}} to do this. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 381: | Line 385: | ||
# Subtract the continuum from the line data | # Subtract the continuum from the line data | ||
contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300' | contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300' | ||
uvcontsub(vis='IRAS16293_Band9.calibrated.tavg.ms',outputvis='IRAS16293_Band9.calibrated.tavg.ms.contsub',fitspec=contspw,fitorder=1) | |||
</source> | </source> | ||
<!-- HAS: is fitorder still 1 if i'm not combining spws?--> | |||
Before we commence with the cleaning we also need to apply the tables we produced in the self calibration steps to the line data. | Before we commence with the cleaning, we also need to apply the tables we produced in the self calibration steps to the line data. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
# Apply final self-calibration to the line data | # Apply final self-calibration to the line data | ||
applycal(vis='IRAS16293_Band9.calibrated.ms.contsub',spwmap=[],spw='', | applycal(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub',spwmap=[],spw='', | ||
gaintable=['cont_pcal2','cont_apcal'],calwt=False,flagbackup=False) | gaintable=['cont_pcal2','cont_apcal'],calwt=False,flagbackup=False) | ||
</source> | </source> | ||
This dataset can produce very large image cubes. To make your work more effective we recommend you | This dataset can produce very large image cubes. To make your work more effective, we recommend you use only a subset of the channels around the line of interest. Also, average every 2 channels since this is the native velocity resolution. If you would like to image the full spws, we recommend splitting them each in half in the tclean calls. The code to image the full spws (each split in half) is provided at the end of this section for reference. We will first focus on the H13CN (J=8-7) line velocity cubes and moment maps. | ||
If you would like to | |||
=== Create a velocity cube for H13CN (J=8-7) === | === Create a velocity cube for H13CN (J=8-7) === | ||
In this section we provide an example for how to create a Doppler corrected velocity cube for one of the interesting lines in the data: H13CN (J=8-7). This is a weaker line in spw 1. | In this section we provide an example for how to create a Doppler corrected velocity cube for one of the interesting lines in the data: H13CN (J=8-7). This is a weaker line in spw 1. | ||
If you need line information, check out the [http://www.cv.nrao.edu/php/splat/ splatalogue] or the 450 micron CSO [http://iopscience.iop.org/0067-0049/132/2/281/ survey] of Orion. | |||
First, determine the velocity range for H13CN 690.552079 GHz with the help of {{plotms_6.6.1}} and the shortest baselines (50m and smaller) to emphasize the extended emission: | |||
First, determine the velocity range for H13CN 690.552079 GHz | |||
with the help of {{plotms_6.6.1}}: | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='IRAS16293_Band9. | plotms(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub', | ||
xaxis='velocity', yaxis='amp',field='', | xaxis='velocity', yaxis='amp',field='', | ||
spw='1', avgtime='1e8',avgscan=True, | spw='1', avgtime='1e8',avgscan=True,iteraxis='field', | ||
transform=True,freqframe='LSRK',restfreq='690.552079GHz', | transform=True,freqframe='LSRK',restfreq='690.552079GHz', | ||
avgchannel='8', | avgchannel='8', ydatacolumn='data', uvrange='<50') | ||
</source> | </source> | ||
There are two lines visible, but since we included the rest frequency for H13CN, the line we want is near 0 km/s for this source. | There are two lines visible (clearest in fields 0 and 2), but since we included the rest frequency for H13CN, the line we want is the weaker one near 0 km/s for this source. | ||
Now do an interactive clean, making clean boxes for the emission. In this case, it is ok to apply the same clean mask to all channels. Stop | Now do an interactive clean, making clean boxes for the emission. In this case, it is ok to apply the same clean mask to all channels. Stop | ||
cleaning when noise-like residuals remain. Number of iterations | cleaning when noise-like residuals remain. Number of iterations | ||
between showing interactive window can be controlled within the | between showing interactive window can be controlled within the | ||
interactive viewer. We used about | interactive viewer. We used about 3000 iterations. Also, note that the weighting scheme is now 'briggsbwtaper' compared to 'briggs' for the aggregate continuum. This is a new weighting scheme available starting in CASA 6.2. See [https://casadocs.readthedocs.io/en/stable/notebooks/synthesis_imaging.html#Imaging-Algorithms Imaging Algorithms] for more information. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
os.system('rm -rf IRAS16293_Band9 | os.system('rm -rf IRAS16293_Band9.H13CN_8_7*') | ||
tclean(vis='IRAS16293_Band9. | tclean(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub', | ||
imagename='IRAS16293_Band9 | imagename='IRAS16293_Band9.H13CN_8_7', | ||
spw='1',field='',phasecenter=3, | spw='1',field='',phasecenter=3, | ||
specmode='cube',outframe='LSRK',restfreq='690.55207GHz', | specmode='cube',outframe='LSRK',restfreq='690.55207GHz', | ||
| Line 491: | Line 443: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
immath('IRAS16293_Band9.H13CN_8_7.image.pbcor', | |||
outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor', | |||
outfile='IRAS16293_Band9 | |||
box='221,160,493,430',expr='IM0') | box='221,160,493,430',expr='IM0') | ||
</source> | </source> | ||
| Line 512: | Line 452: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.fits') | |||
fitsimage='IRAS16293_Band9 | |||
</source> | </source> | ||
=== Moment Maps for H13CN (J=8-7) === | === Moment Maps for H13CN (J=8-7) === | ||
To proceed with the moment maps generation, you will need to determine the channel range and pixel range for inclusion for higher order moments. | To proceed with the moment maps generation, you will need to determine the channel range and pixel range for inclusion for higher order moments, which you can do by inspecting the cube in CARTA. You want to choose only the channels that you are sure have real emission. | ||
Generate the moment 0 map using the {{immoments_6.6.1}} task. You will see warnings about the convolving kernel. This is ok. | |||
Generate the moment 0 map. You will see warnings about the convolving kernel. This is ok. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
os.system('rm -rf IRAS16293_Band9 | os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0') | ||
immoments(imagename='IRAS16293_Band9 | immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',moments=[0], | ||
outfile='IRAS16293_Band9 | outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0', | ||
chans='10~44') | chans='10~44') | ||
</source> | </source> | ||
| Line 552: | Line 476: | ||
[[File:IRAS16293_Band9.fixed.H13CN_8_7.Negative_mom1_6.5.4.png|200px|thumb|right|'''Fig. 10.''' Negative part of the moment 1.]] | [[File:IRAS16293_Band9.fixed.H13CN_8_7.Negative_mom1_6.5.4.png|200px|thumb|right|'''Fig. 10.''' Negative part of the moment 1.]] | ||
One cannot make higher order moments that simultaneously include | One cannot make higher order moments that simultaneously include | ||
both positive and negative emission | both positive and negative emission, so we must separate them. Also, we need to restrict the higher order moments to regions | ||
of real signal. One way to do this is to use your clean mask to limit where | of real signal. One way to do this is to use your clean mask to limit where | ||
we calculate the | we calculate the higher order moments. | ||
Here we demonstrate how to use the clean mask for this | |||
purpose. We must first subimage it to match the primary beam corrected image. | purpose. We must first subimage it to match the primary beam corrected image. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
immath(imagename='IRAS16293_Band9 | immath(imagename='IRAS16293_Band9.H13CN_8_7.mask', | ||
mode='evalexpr',expr='IM0',box='221,160,493,430', | mode='evalexpr',expr='IM0',box='221,160,493,430', | ||
outfile='IRAS16293_Band9 | outfile='IRAS16293_Band9.H13CN_8_7.mask.subim') | ||
</source> | </source> | ||
Producing the first and second moments in two steps, focusing first on the positive emission, | We also limit the pixels used to those with line emission greater than approximately 5sigma | ||
(5 times the rms noise value of the image). Producing the first and second moments in two steps, focusing first on the positive emission, | |||
and second on the negative (absorption) features: | and second on the negative (absorption) features: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
os.system('rm -rf IRAS16293_Band9 | os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive*') | ||
immoments(imagename='IRAS16293_Band9 | immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',moments=[1,2], | ||
outfile='IRAS16293_Band9 | outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive', | ||
mask='IRAS16293_Band9 | mask='IRAS16293_Band9.H13CN_8_7.mask.subim', | ||
chans='10~44',includepix=[0.4,100]) | chans='10~44',includepix=[0.4,100]) | ||
os.system('rm -rf IRAS16293_Band9 | os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative*') | ||
immoments(imagename='IRAS16293_Band9 | immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',moments=[1,2], | ||
outfile='IRAS16293_Band9 | outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative', | ||
mask='IRAS16293_Band9 | mask='IRAS16293_Band9.H13CN_8_7.mask.subim', | ||
chans='10~44',includepix=[-100,-0.4]) | chans='10~44',includepix=[-100,-0.4]) | ||
</source> | </source> | ||
Use CARTA to visualize the maps. | Use CARTA to visualize the maps. | ||
And finally export the result to FITS format for further analysis: | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.fits') | |||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.fits') | |||
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0.fits') | |||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0.fits') | |||
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_coord.fits') | |||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_coord', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_coord.fits') | |||
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_coord.fits') | |||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_coord', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_coord.fits') | |||
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_dispersion_coord.fits') | |||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_dispersion_coord', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_dispersion_coord.fits') | |||
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_dispersion_coord.fits') | |||
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_dispersion_coord', | |||
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_dispersion_coord.fits') | |||
</source> | |||
=== Clean each spw === | |||
Some notes for this section: the threshold was chosen after doing a couple of smaller test images. This is about 3 sigma in channels with weak line emission. The mask was also chosen from a few test images. By setting the mask like this, there is no need to do interactive cleaning. Note however, that this clean mask does not do a good job on the extended emission of CO J=6-5. You may wish to experiment with the {{automask_6.6.1}} algorithm in {{tclean_6.6.1}}. | |||
You will see a warning that no rest frequency has been specified. You may wish to modify the rest frequency for each spw in order to have more accurate velocity labeling. A good reference for finding rest frequencies is [http://www.cv.nrao.edu/php/splat/ splatalogue]. Another good reference for Band 9 specifically is the 450 micron CSO survey of Orion: http://iopscience.iop.org/0067-0049/132/2/281/ | |||
If you would like to look at the initial cube and where the masks are located, use the {{tclean_6.6.1}} call in the loop to create an interactive clean session for one spw by setting 'interactive=True' and replacing 'str(i)' with the spw number you wish to clean. This will pop up an initial {{tclean_6.6.1}} window as before. Click through the cube channels using the arrow buttons in the 'Animator' section of the window. You can do one or more rounds of interactive cleaning, and then click the blue arrow under 'Next action' to let {{tclean_6.6.1}} finish non-interactively. Note that every time {{tclean_6.6.1}} needs to create a new cube for interactive viewing, it will take a while! | |||
We first clean the first half of each spw. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf *spw*a.*') | |||
spw=[0,1,2,3] | |||
for i in spw: | |||
tclean(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub', | |||
imagename='IRAS16293_Band9.spw'+str(i)+'a', | |||
spw=str(i),field='',phasecenter=3, | |||
specmode='cube',outframe='LSRK', | |||
nchan=950,start=20,width=2, | |||
gridder='mosaic',mosweight=True, | |||
imsize=720,cell='0.028arcsec', | |||
interactive=False,restoringbeam='common', | |||
mask=['circle[[16h32m22.61s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.86s,-24d28m36.57s ], 1.0arcsec]'], | |||
weighting='briggsbwtaper',robust=0.5,pbcor=True, | |||
niter=500, threshold='220.0mJy') | |||
</source> | </source> | ||
Next clean the 2nd half of each spw. | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
os.system('rm -rf *spw*b.*') | |||
spw=[0,1,2,3] | |||
for i in spw: | |||
tclean(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub', | |||
imagename='IRAS16293_Band9.spw'+str(i)+'b', | |||
spw=str(i),field='',phasecenter=3, | |||
specmode='cube',outframe='LSRK', | |||
nchan=950,start=1918,width=2, | |||
gridder='mosaic',mosweight=True, | |||
imsize=720,cell='0.028arcsec', | |||
interactive=False,restoringbeam='common', | |||
mask=['circle[[16h32m22.61s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.86s,-24d28m36.57s ], 1.0arcsec]'], | |||
weighting='briggsbwtaper',robust=0.5,pbcor=True, | |||
niter=500, threshold='220.0mJy') | |||
</source> | </source> | ||
Create subset images of the line cubes to center on the sources and export to FITS: | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
spw=[0,1,2,3] | |||
for i in spw: | |||
immath('IRAS16293_Band9.spw'+str(i)+'a.image.pbcor', | |||
outfile='IRAS16293_Band9.spw'+str(i)+'a.image.subim.pbcor', | |||
box='221,160,493,430',expr='IM0') | |||
spw=[0,1,2,3] | |||
for i in spw: | |||
immath('IRAS16293_Band9.spw'+str(i)+'b.image.pbcor', | |||
outfile='IRAS16293_Band9.spw'+str(i)+'b.image.subim.pbcor', | |||
box='221,160,493,430',expr='IM0') | |||
spw=[0,1,2,3] | |||
exportfits(imagename='IRAS16293_Band9. | for i in spw: | ||
exportfits(imagename='IRAS16293_Band9.spw'+str(i)+'a.image.subim.pbcor', | |||
fitsimage='IRAS16293_Band9.spw'+str(i)+'a.image.subim.pbcor.fits') | |||
exportfits(imagename='IRAS16293_Band9. | |||
spw=[0,1,2,3] | |||
for i in spw: | |||
exportfits(imagename='IRAS16293_Band9.spw'+str(i)+'b.image.subim.pbcor', | |||
fitsimage='IRAS16293_Band9.spw'+str(i)+'b.image.subim.pbcor.fits') | |||
</source> | </source> | ||
Latest revision as of 22:40, 20 December 2024
Most recently updated for CASA Version 6.6.1 using Python 3.8
This guide features CARTA, the “Cube Analysis and Rendering Tool for Astronomy,” which is the new NRAO visualization tool for images and cubes. The CASA viewer (imview) has not been maintained for a few years and will be removed from future versions of CASA. We strongly recommend using CARTA, as it provides a much more efficient, stable, and feature rich user experience. A comparison of the CASA viewer and CARTA, as well as instructions on how to use CARTA at NRAO, is provided in the CARTA section of the CASA docs.
Overview and Setup
This section of the CASAguide continues from IRAS16293 Band9 - Calibration for CASA 6.6.1 . If you completed the calibration section, then you already have the final calibrated dataset. If you just started to work on this IRAS16293 Band 9 data, you can download the fully calibrated dataset IRAS16293_Band9.calibrated.tavg.ms.tgz here, as well as a mask file IRAS16293_Band9.cont.mask to be used for the selfcal and continuum imaging section.
This CASAguide covers imaging of both continuum and spectral lines of IRAS16293 Band 9. Details of the ALMA observations are provided at IRAS16293 Band9. This guide requires CASA 6.6.1. Please confirm your version before proceeding.
# In CASA
import casalith
version = casalith.version_string()
print ("You are using " + version)
if (version < '6.6.1'):
print ("YOUR VERSION OF CASA IS TOO OLD FOR THIS GUIDE.")
print ("PLEASE UPDATE IT BEFORE PROCEEDING.")
else:
print ("Your version of CASA is appropriate for this guide.")
This CASAguide also uses Analysis Utilities (or analysisUtils for short), which is a small set of Python scripts that provide a number of analysis and plotting utilities for ALMA data reduction. They are very easy to install (just download and untar). See here for a full description and download instructions. Analysis Utilities are updated frequently so if its been a while since you installed it, its probably worth doing it again. If you are at an ALMA site or ARC, the analysis utilities are already installed and up to date.
Before we start with the imaging itself, do a listobs to the calibrated dataset, so you can check that everything is ok. Make sure there are 4 observations, 7 fields, and 4 spectral windows (spws).
# In CASA
# listobs to check data
listobs(vis='IRAS16293_Band9.calibrated.ms',
listfile='IRAS16293_Band9.calibrated.tavg.ms.listobs',verbose=False)
You should see the next information in the .listobs file.
Telescope Observation Date Observer Project ALMA [ 4.84128e+09, 4.84129e+09]dgarcia uid://A002/X3cd6b2/X53 ALMA [ 4.84135e+09, 4.84136e+09]dgarcia uid://A002/X3cd6b2/X53 ALMA [ 4.84137e+09, 4.84137e+09]dgarcia uid://A002/X3cd6b2/X53 ALMA [ 4.84137e+09, 4.84138e+09]dgarcia uid://A002/X3cd6b2/X53 Data records: 52996 Total elapsed time = 98524.9 seconds Observed from 16-Apr-2012/08:27:40.4 to 17-Apr-2012/11:49:45.3 (UTC) Fields: 7 ID Code Name RA Decl Epoch SrcId nRows 0 none IRAS16293-2422-a 16:32:22.992000 -24.28.36.00000 J2000 0 7812 1 none IRAS16293-2422-a 16:32:22.479253 -24.28.36.00000 J2000 0 7821 2 none IRAS16293-2422-a 16:32:22.735626 -24.28.36.00000 J2000 0 7821 3 none IRAS16293-2422-a 16:32:22.735626 -24.28.32.50000 J2000 0 7821 4 none IRAS16293-2422-a 16:32:22.479253 -24.28.29.00000 J2000 0 7439 5 none IRAS16293-2422-a 16:32:22.735626 -24.28.29.00000 J2000 0 7141 6 none IRAS16293-2422-a 16:32:22.992000 -24.28.29.00000 J2000 0 7141 Spectral Windows: (4 unique spectral windows and 1 unique polarization setups) SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) BBC Num Corrs 0 ALMA_RB_09#BB_1#SW-01#FULL_RES 3840 TOPO 703312.744 488.281 1875000.0 704250.0000 1 XX YY 1 ALMA_RB_09#BB_2#SW-01#FULL_RES 3840 TOPO 692237.256 -488.281 1875000.0 691300.0000 2 XX YY 2 ALMA_RB_09#BB_3#SW-01#FULL_RES 3840 TOPO 690437.256 -488.281 1875000.0 689500.0000 3 XX YY 3 ALMA_RB_09#BB_4#SW-01#FULL_RES 3840 TOPO 688437.256 -488.281 1875000.0 687500.0000 4 XX YY Antennas: 13 'name'='station' ID= 0-3: 'DA41'='A003', 'DA43'='A075', 'DA47'='A026', 'DV02'='A077', ID= 4-8: 'DV03'='A137', 'DV07'='A076', 'DV09'='A046', 'DV10'='A071', ID= 9-13: 'DV12'='A011', 'DV13'='A072', 'DV14'='A025', 'DV17'='A138', ID= 14-14: 'DV18'='A053'
Continuum Emission Imaging
As shown in Figure 2 of IRAS16293 Band9, these observations have a 7 pointing mosaic, which you can visualize with the next command (see also Figure 1).
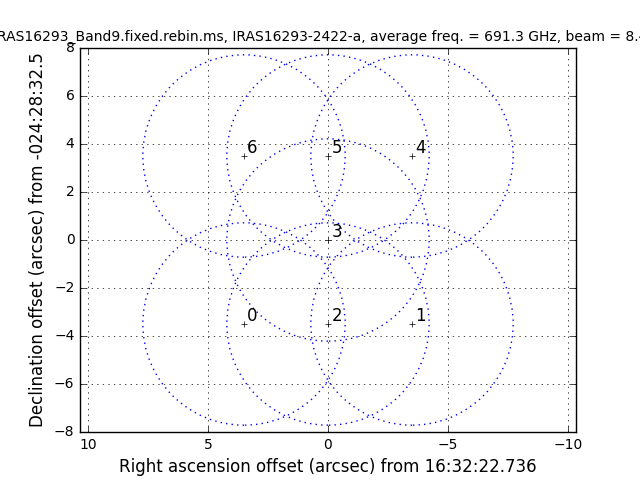
# In CASA
# Make a plot of the mosaic pattern with field ids identified
au.plotmosaic(vis='IRAS16293_Band9.calibrated.tavg.ms',sourceid='0',
doplot=True,figfile='mosaic_pattern.png')
You might wonder which of these pointings or fields have stronger line emission. You can determine this by running the next command. We set spw='1' which contains CO (6-5), and you can check all the plots for all the fields by just hitting the arrow for "next". Also see Figure 2 for an example of the output.
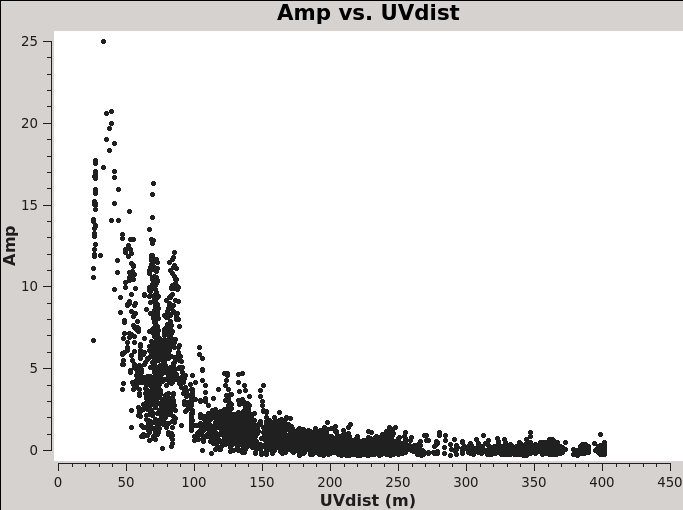
# In CASA
# See which mosaic fields have the brightest line emission
plotms(vis='IRAS16293_Band9.calibrated.tavg.ms',
field='',xaxis='uvdist', yaxis='amp',
spw='1', avgchannel='3840',coloraxis='field',
iteraxis='field',yselfscale=True,showgui=True)
Another way to visualize the uv-amp distribution is with the next plotms. This will load all the fields for pointing into the plot, and you can change interactively between the spws using the appropriate button in plotms. See Figure 3 for a plot of spw 0. Remember you can use the locate and zoom features to find more information about each plotted point.
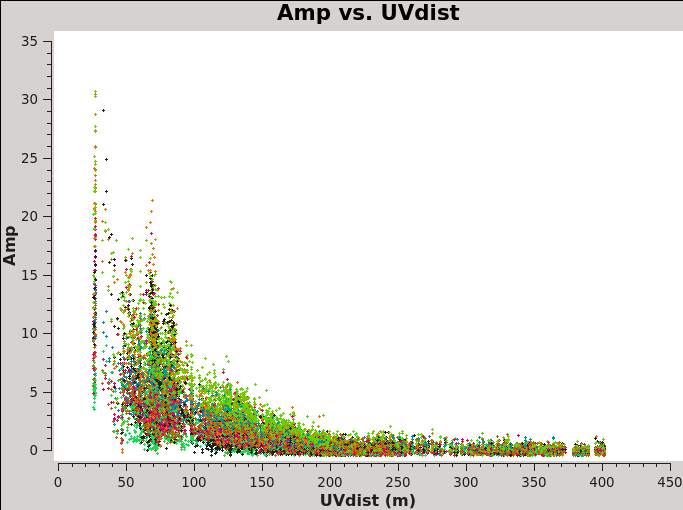
# In CASA
# Iterate over spws all fields
plotms(vis='IRAS16293_Band9.calibrated.tavg.ms',
field='',xaxis='uvdist', yaxis='amp',
spw='', avgchannel='3840',coloraxis='field',
iteraxis='spw',showgui=True)
In order to properly measure the amount of CO(6-5) emission, we need to note the channels that do not present line emission. We will use these channels to image the continuum and subtract the continuum prior to line imaging. To identify them we can plot the amplitude vs frequency for all the spectral windows.
Figure 4 shows the output from the next command for spw 0 (field 3). Note: If you are using the calibrated data from the science portal, or if you calibrated the raw data yourself following the steps in the IRAS16293_Band9_-_Calibration CASA Guide, you now have a set of data which were split, so the calibrated data now reside in the "data" column of the new measurement set. Therefore the following call to plotms must specify ydatacolumn = "data". If you never performed the split, the calibrated data would have been accessed from the "corrected" column of the old, unsplit, measurement set. Those who have used previous versions of CASA should note that in those older versions, plotms would default to using the "data" column if there were no data in the "corrected" column and one specified ydatacolumn="corrected".
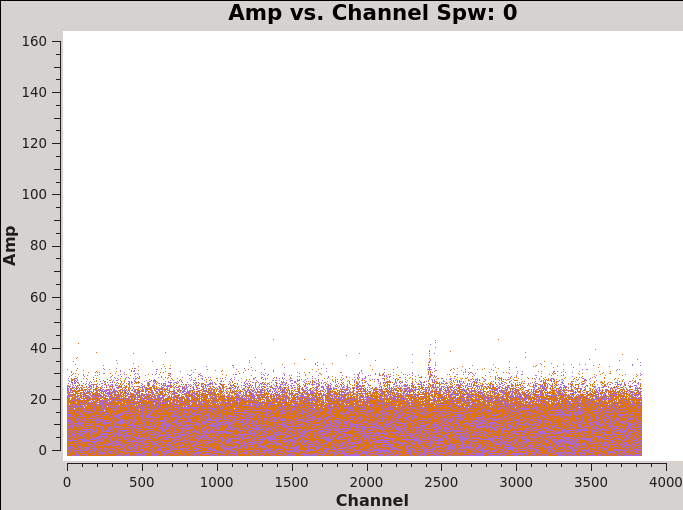
# In CASA
# Select Channels for uvcontsub. Average every 8 channels for
# increased S/N.
plotms(vis='IRAS16293_Band9.calibrated.tavg.ms',
xaxis='channel', yaxis='amp',field='3',
spw='', avgtime='1e8',avgscan=True,coloraxis='corr',
avgchannel='',iteraxis='spw',ydatacolumn='data',
xselfscale=True,yselfscale=True,showgui=True)
Using the plots from the previous command you can select a block of channels that are free of strong line emission. To do this in the most rigorous way, you would first need to make dirty image cubes and then examine the spectra on and near the continuum sources, because many more weaker lines will become apparent in the cubes. See Image_Continuum#Create_an_Averaged_Continuum_MS for more detailed instructions and commands to do this. Here we provide you with the channel selection we used for continuum emission.
The next flagmanager command will save the flags state in case you might need to re-do the steps from this point.
# In CASA
flagmanager(vis='IRAS16293_Band9.calibrated.tavg.ms',mode='save',versionname='Original')
Now we will flag the channels identified to have line emission, so we will be ready to proceed with the continuum imaging.
# In CASA
# flagging the line channels
flagdata(vis='IRAS16293_Band9.calibrated.tavg.ms',
spw='0:1700~2500,1:400~500;1100~1900;3000~3450,2:1700~2200;3100~3839,3:0~800;1600~2900;3300~3839',action='apply', flagbackup=False)
To help tclean to be faster, you can average 200 channels, as you do not need to have a high spectral resolution for continuum imaging. See Image_Continuum#Create_an_Averaged_Continuum_MS for more information on choosing the ideal width to average over while avoiding bandwidth smearing.
# In CASA
split(vis='IRAS16293_Band9.calibrated.tavg.ms',
outputvis='IRAS16293_Band9.calibrated.tavg.chavg.ms',field='',
datacolumn='data',width=200,spw='0~3:20~3819')
Selfcal Round 1
Now we proceed with the cleaning itself. For the tclean task you will need to choose the right cell size, map size, etc. Below are the parameters that were used in this CASAguide. For further discussion on imaging parameters, see Image_Continuum#Image_Parameters. Note that the command will be interactive and you will have to set the cleaning masks. For a basic guide to cleaning, see First_Look_at_Imaging.
Since we will be self-calibrating this dataset, we will be conservative in this first tclean run. Mask only the brightest emission, and only run for one or two major cycles to create a first, initial model of the source emission. We have provided a mask in IRAS16293_Band9.calibrated.cont.mask.tgz, which you should untar with tar -xvzf if you wish to use it. If you would rather create your own mask, delete or change the name of the provided mask if you have downloaded it.
Using tclean with gridder = 'mosaic' allows us to set an additional parameter mosweight with a default of True. When mosweight = True, the gridder weights each field in the mosaic independently. The mosweight parameter is particularly important for mosaics with non-uniform sensitivity, with rectangular shapes, or when using more uniform values of robust Briggs weighting. For more information on mosweight, please see the tclean documentation.
Note that starting in CASA 6.2, the beam-fitting algorithm has been updated.
# In CASA
# cleaning 1: do not delete IRAS16293_Band9.calibrated.cont.mask if you want to use the provided mask
for ext in ['.image','.model','.image.pbcor','.psf','.residual','.pb','.sumwt','.weight']:
rmtables('IRAS16293_Band9.calibrated.cont'+ext)
tclean(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',
imagename='IRAS16293_Band9.calibrated.cont',
spw='0~3',field='',phasecenter=3,
specmode='mfs',gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=True,deconvolver='hogbom',nterms=1,
weighting='briggs',robust=0.5,savemodel='modelcolumn',
niter=1000, threshold='75.0mJy')
It is important to check if all fields have some emission in the model data column. If there is no emisison, or if it is very weak, you would want to exclude those fields from the gaincal below. Because the bright Source B is within the primary beam of all the observed fields, all fields have strong emission present in the image model. In Figure 5 you can see an example for the output from next command. If there was no signal for one of the fields, you would see values at 0 amplitude.
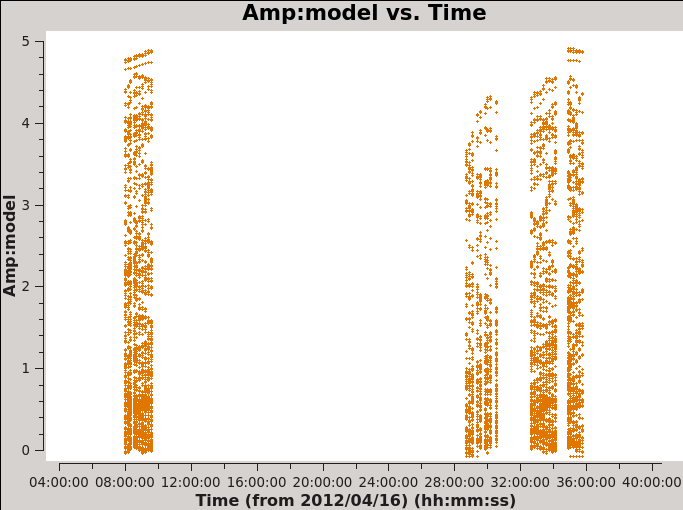
# In CASA
# checking the model data for all the fields
plotms(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',
xaxis='time', yaxis='amp',field='',iteraxis='field',
spw='', avgchannel='4000',coloraxis='field',showgui=True,
ydatacolumn='model',xselfscale=True,yselfscale=True)
Now we proceed with gaincal to perform self calibration (for phases) on the data. Even with minsnr=3.0 there will be some failed solutions. For more information on self-calibration see First_Look_at_Self_Calibration.
# In CASA
# get one scan per field
os.system('rm -rf cont_pcal1')
gaincal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',caltable='cont_pcal1',gaintype='T',
refant='DV14',calmode='p',combine='',spw='',field='',
solint='inf',minsnr=3.0,minblperant=4)
The solutions for five antennas are shown in Figure 6, and are produced with the next plotms. Be sure to check all the antennas. Look for apparent large corrections or wrapping. Remember these are the corrections, not the data. Note any antennas that seem concerning.
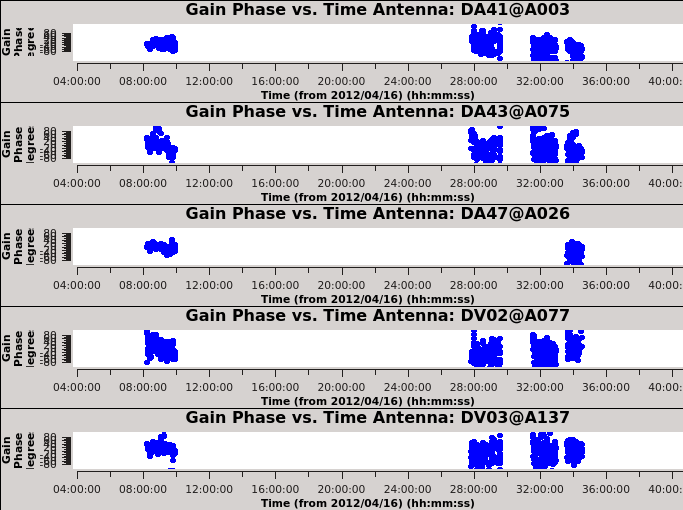
# In CASA
plotms(vis='cont_pcal1',xaxis='time',yaxis='phase',
spw='',iteraxis='antenna',gridrows=5,gridcols=1,plotrange=[0,0,-80,80],
antenna='',timerange='',showgui=True)
# The phases are remarkably stable for Band 9!
Before applying the calibrations to the data, we will again save the flags state in case we want to re-do the steps from this point. To restore the flags, set 'mode='restore'.
# In CASA
flagmanager(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',mode='save',versionname='Original')
We then apply the table to the data. We will check the solutions by cleaning the data again and comparing the self-calibrated and non-self-calibrated data.
# In CASA
# application of the table
applycal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',spwmap=[],spw='',
gaintable=['cont_pcal1'],calwt=False,flagbackup=False)
Selfcal Round 2
Now you are ready for the second cleaning. For this one we are using the mask resulting from the previous cleaning. In the next blocks of commands we will repeat this process (clean, carta, gaincal, plotms, applycal, clean, etc.) to improve the overall quality of the image. You can add to your clean mask as you go, but be careful to avoid adding sidelobe emission as real structure.
# In CASA
# cleaning 2
os.system('rm -rf IRAS16293_Band9.calibrated.cont_p1.*')
tclean(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',
imagename='IRAS16293_Band9.calibrated.cont_p1',
mask='IRAS16293_Band9.calibrated.cont.mask',
spw='0~3',field='',phasecenter=3,
specmode='mfs',gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=True,deconvolver='hogbom',nterms=1,
weighting='briggs',robust=0.5,savemodel='modelcolumn',
niter=1000, threshold='50.0mJy')
After tclean has finished, you should now open the image in CARTA. If using NRAO machines, you can open a new terminal tab, cd to the working directory, then type:
#in terminal
carta --no_browser
Copy the output URL into a browser to view your CARTA session. Select and load "IRAS16293_Band9.calibrated.cont_p1.image".
The image "IRAS16293_Band9.calibrated.cont_p1.image" should have 2 very clearly defined regions of emission, moreso than the first clean run "IRAS16293_Band9.calibrated.cont.image," following self-calibration.
Second gaincal step for self calibration:
# In CASA
# second self-cal
os.system('rm -rf cont_pcal2')
gaincal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',caltable='cont_pcal2',gaintype='T',
refant='DV14',calmode='p',combine='',spw='',field='',
solint='inf',minsnr=3.0,minblperant=4)
#Checking the table
plotms(vis='cont_pcal2',xaxis='time',yaxis='phase',
spw='',iteraxis='antenna',gridrows=5,gridcols=1,plotrange=[0,0,-80,80],
antenna='',timerange='',showgui=True)
# Applying results to the data
applycal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',spwmap=[],spw='',
gaintable=['cont_pcal2'],calwt=False,flagbackup=False)
Selfcal Round 3
We continue with the next cleaning.
# In CASA
# Cleaning 3
os.system('rm -rf IRAS16293_Band9.calibrated.cont_p2*')
tclean(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',
imagename='IRAS16293_Band9.calibrated.cont_p2',
mask='IRAS16293_Band9.calibrated.cont_p1.mask',
spw='0~3',field='',phasecenter=3,
specmode='mfs',gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=True,deconvolver='hogbom',nterms=1,
weighting='briggs',robust=0.5,savemodel='modelcolumn',
niter=1000, threshold='40.0mJy')
Open "IRAS16293_Band9.calibrated.cont_p2.image" in CARTA. There is very little change; the bold could try another iteration of phase self-cal using a shorter solint. For now, let's move on to amplitude.
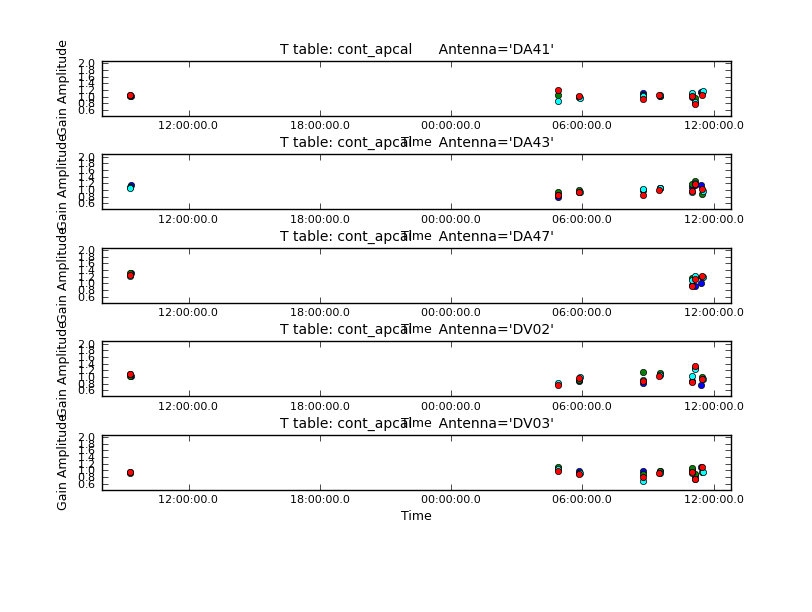
Next iteration with gaincal, now using amplitude and phase for "calmode".
# In CASA
# For amplitude self-cal we only want to get one solution per spw, per dataset
os.system('rm -rf cont_apcal')
gaincal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',
caltable='cont_apcal',gaintype='T',refant='DV14',
calmode='ap',combine='scan,field',spw='',field='',
solint='inf',minsnr=3.0,minblperant=4,gaintable='cont_pcal2')
# ploting the table
plotms(vis='cont_apcal',xaxis='time',yaxis='amp',
spw='',iteraxis='antenna',gridrows=5,gridcols=1,
plotrange=[0,0,0.5,2],antenna='',timerange='',
showgui=True)
# and applying to the data
applycal(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',spwmap=[],
spw='',gaintable=['cont_pcal2','cont_apcal'],calwt=False,
flagbackup=False)
# A large correction is present for the 2nd dataset. This
# amplitude error was causing many of the artifacts noticeable
# in the previous images.
Final Aggregate Continuum Image
Continuing the final cleaning step. We will re-use the mask from the cont_p2 image, and also set pbcor=True to output a pb-corrected image.
# In CASA
# Cleaning 4
os.system('rm -rf IRAS16293_Band9.calibrated.cont_ap.*')
tclean(vis='IRAS16293_Band9.calibrated.tavg.chavg.ms',
imagename='IRAS16293_Band9.calibrated.cont_ap',
spw='0~3',field='',phasecenter=3,
specmode='mfs',gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=True,deconvolver='hogbom',nterms=1,
mask='IRAS16293_Band9.calibrated.cont_p2.mask',
weighting='briggs',robust=0.5,savemodel='modelcolumn',
niter=1000, threshold='40.0mJy',pbcor=True)
Open "IRAS16293_Band9.calibrated.cont_ap.image" in CARTA.
We will use immath to cut out a sub-region of the cube because the area covered is large compared to the area of interest, and we want to keep the file size of the result as small as possible.
# in CASA
immath('IRAS16293_Band9.calibrated.cont_ap.image.pbcor',outfile='IRAS16293_Band9.calibrated.cont_ap.image.subim.pbcor',
box='221,160,493,430',expr='IM0')
Note that the default outfile is immath_results.im, if you do not specify an outfile name. Check the image in CARTA.
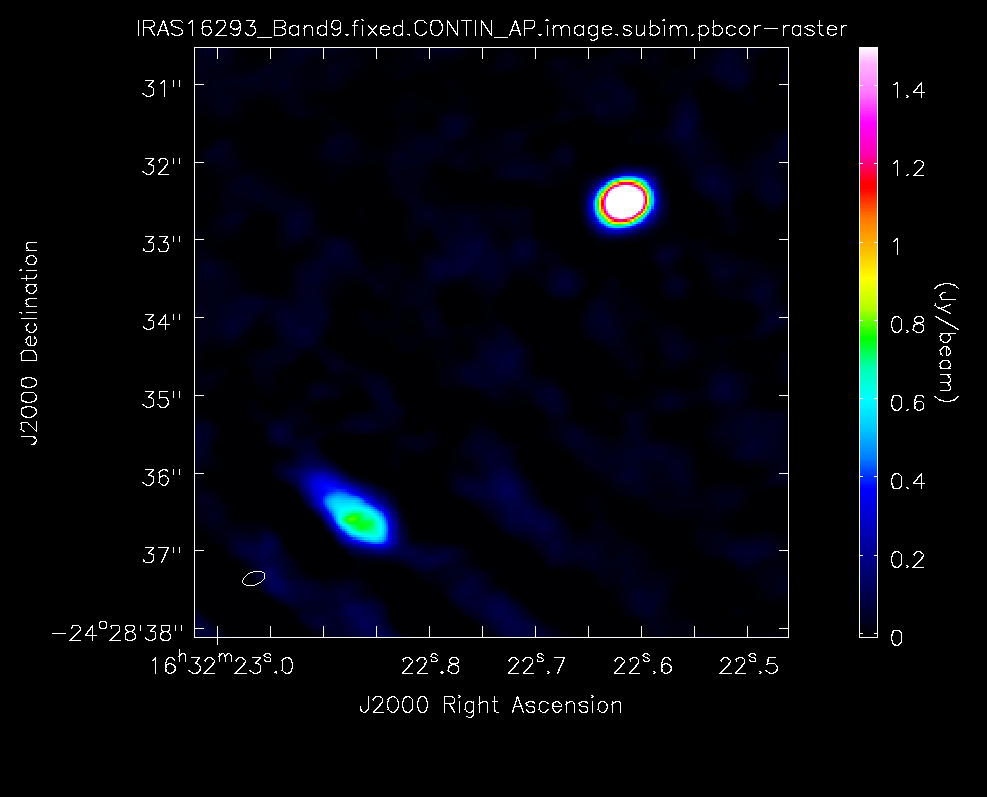
In Figure 7 we show the final image of the self-calibration process. Source A is the weaker source to the SE and Source B is the stronger source to the NW. The rms noise is about 18 mJy/beam, and the restoring beam is 0.30 x 0.16 arcsec, PA=-70 deg with robust=0.5 weighting. The peak intensities for the two components are about 3.07 (B) and 1.02 (A) Jy/beam, while the flux densities are about 10 (B) and 8.3 (A) Jy.
Now you can export your final image in FITS format, so you can analyze it in other software packages if desired.
# In CASA
exportfits(imagename='IRAS16293_Band9.calibrated.cont_ap.image.subim.pbcor',
fitsimage='IRAS16293_Band9.calibrated.cont_ap.image.subim.pbcor.fits')
Spectral Line Imaging
We need to restore the line channels we flagged when creating the continuum.
# In CASA
# Restore the flags made for the continuum imaging
flagmanager(vis='IRAS16293_Band9.calibrated.tavg.ms',mode='restore',versionname='Original')
Then we need to subtract the continuum, using the line free channels. The line free channels are just the inverse of the line channels identified earlier. We will use uvcontsub to do this.
# In CASA
# Subtract the continuum from the line data
contspw='0:20~1700;2500~3820,1:20~400;500~1100;1900~3000;3450~3800,2:20~1700;2200~3100,3:800~1600;2900~3300'
uvcontsub(vis='IRAS16293_Band9.calibrated.tavg.ms',outputvis='IRAS16293_Band9.calibrated.tavg.ms.contsub',fitspec=contspw,fitorder=1)
Before we commence with the cleaning, we also need to apply the tables we produced in the self calibration steps to the line data.
# In CASA
# Apply final self-calibration to the line data
applycal(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub',spwmap=[],spw='',
gaintable=['cont_pcal2','cont_apcal'],calwt=False,flagbackup=False)
This dataset can produce very large image cubes. To make your work more effective, we recommend you use only a subset of the channels around the line of interest. Also, average every 2 channels since this is the native velocity resolution. If you would like to image the full spws, we recommend splitting them each in half in the tclean calls. The code to image the full spws (each split in half) is provided at the end of this section for reference. We will first focus on the H13CN (J=8-7) line velocity cubes and moment maps.
Create a velocity cube for H13CN (J=8-7)
In this section we provide an example for how to create a Doppler corrected velocity cube for one of the interesting lines in the data: H13CN (J=8-7). This is a weaker line in spw 1. If you need line information, check out the splatalogue or the 450 micron CSO survey of Orion.
First, determine the velocity range for H13CN 690.552079 GHz with the help of plotms and the shortest baselines (50m and smaller) to emphasize the extended emission:
# In CASA
plotms(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub',
xaxis='velocity', yaxis='amp',field='',
spw='1', avgtime='1e8',avgscan=True,iteraxis='field',
transform=True,freqframe='LSRK',restfreq='690.552079GHz',
avgchannel='8', ydatacolumn='data', uvrange='<50')
There are two lines visible (clearest in fields 0 and 2), but since we included the rest frequency for H13CN, the line we want is the weaker one near 0 km/s for this source.
Now do an interactive clean, making clean boxes for the emission. In this case, it is ok to apply the same clean mask to all channels. Stop cleaning when noise-like residuals remain. Number of iterations between showing interactive window can be controlled within the interactive viewer. We used about 3000 iterations. Also, note that the weighting scheme is now 'briggsbwtaper' compared to 'briggs' for the aggregate continuum. This is a new weighting scheme available starting in CASA 6.2. See Imaging Algorithms for more information.
# In CASA
os.system('rm -rf IRAS16293_Band9.H13CN_8_7*')
tclean(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub',
imagename='IRAS16293_Band9.H13CN_8_7',
spw='1',field='',phasecenter=3,
specmode='cube',outframe='LSRK',restfreq='690.55207GHz',
nchan=60,start='-10km/s',width='0.5km/s',
gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=True,
weighting='briggsbwtaper',robust=0.5,
restoringbeam='common',
niter=10000, threshold='220.0mJy',pbcor=True)
Create subset images of the line cubes to center on the sources:
# In CASA
immath('IRAS16293_Band9.H13CN_8_7.image.pbcor',
outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',
box='221,160,493,430',expr='IM0')
To enable analysis in other software packages, you can export the data from CASA to FITS format using exportfits.
# In CASA
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.fits')
Moment Maps for H13CN (J=8-7)
To proceed with the moment maps generation, you will need to determine the channel range and pixel range for inclusion for higher order moments, which you can do by inspecting the cube in CARTA. You want to choose only the channels that you are sure have real emission.
Generate the moment 0 map using the immoments task. You will see warnings about the convolving kernel. This is ok.
# In CASA
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0')
immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',moments=[0],
outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0',
chans='10~44')
Figure 8 shows the moment 0 map. Use CARTA to open the image.
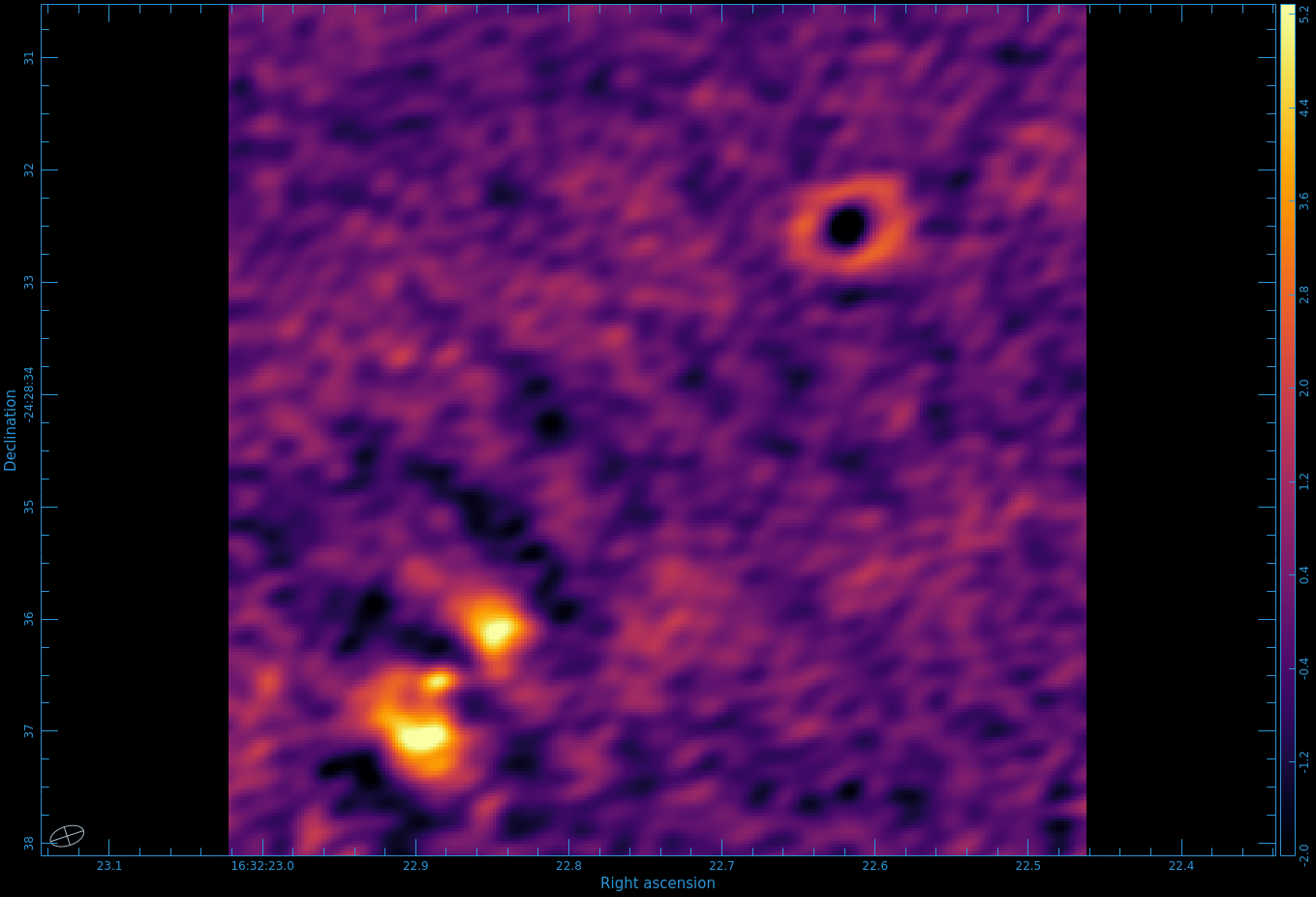
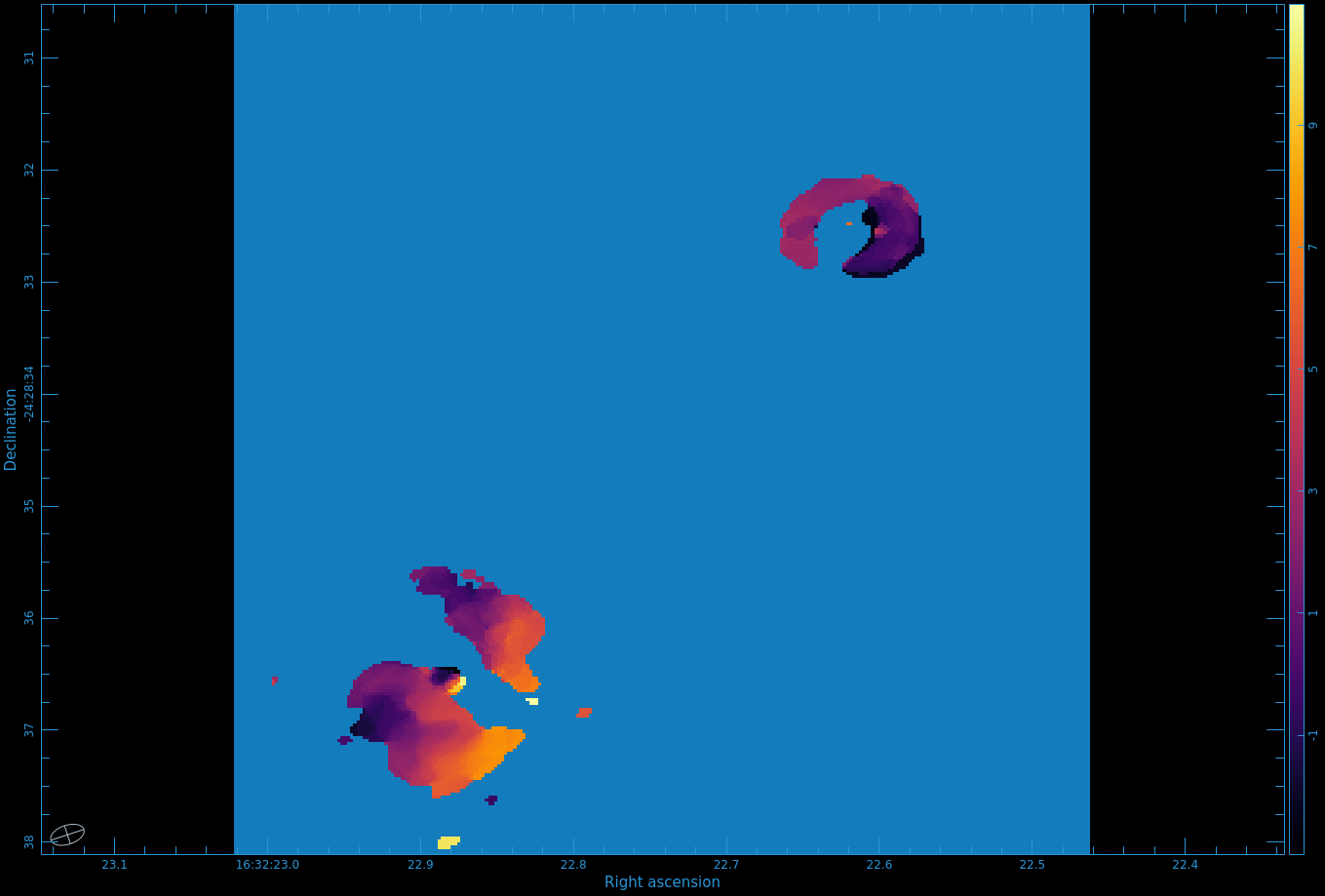
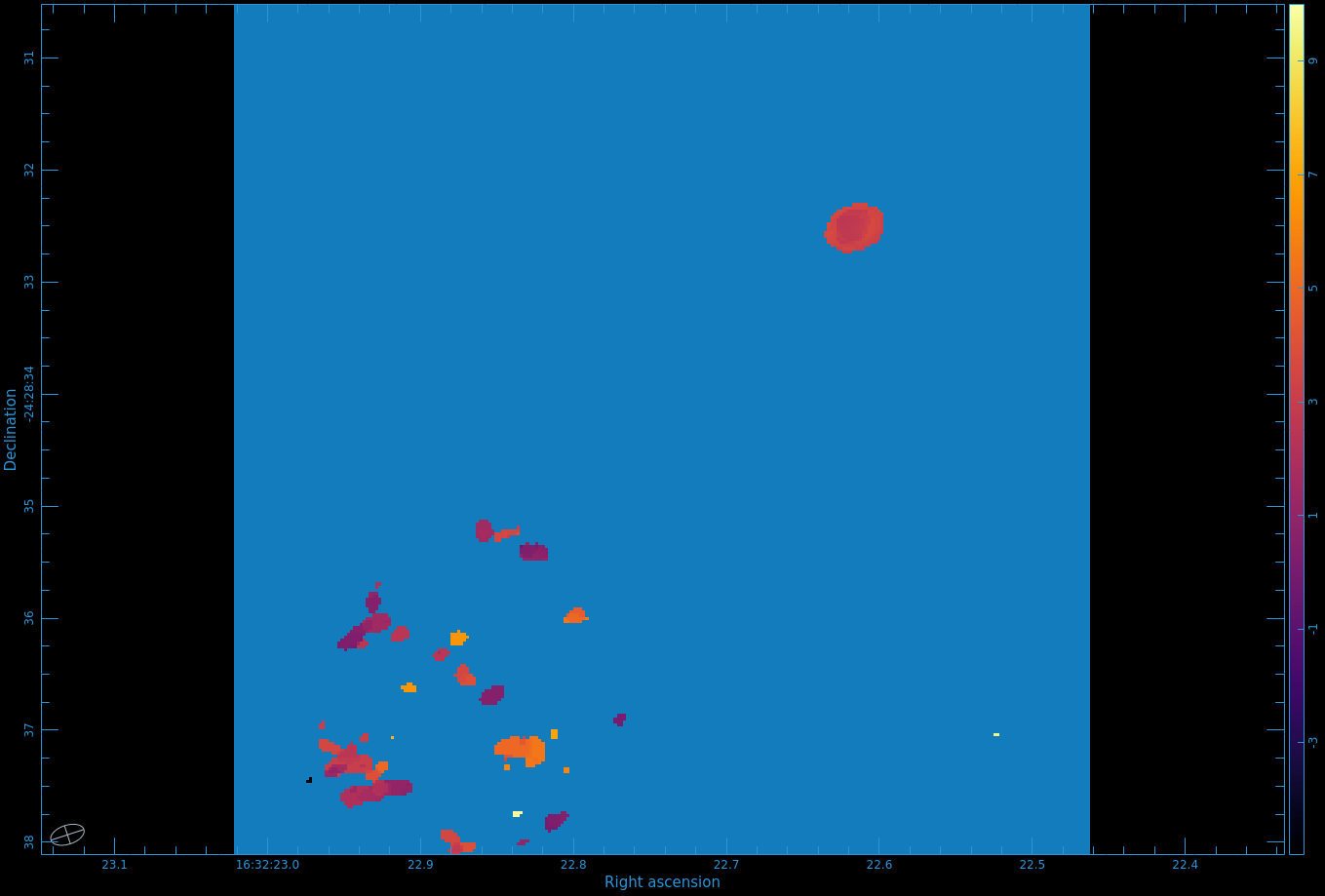
One cannot make higher order moments that simultaneously include both positive and negative emission, so we must separate them. Also, we need to restrict the higher order moments to regions of real signal. One way to do this is to use your clean mask to limit where we calculate the higher order moments.
Here we demonstrate how to use the clean mask for this purpose. We must first subimage it to match the primary beam corrected image.
# In CASA
immath(imagename='IRAS16293_Band9.H13CN_8_7.mask',
mode='evalexpr',expr='IM0',box='221,160,493,430',
outfile='IRAS16293_Band9.H13CN_8_7.mask.subim')
We also limit the pixels used to those with line emission greater than approximately 5sigma (5 times the rms noise value of the image). Producing the first and second moments in two steps, focusing first on the positive emission, and second on the negative (absorption) features:
# In CASA
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive*')
immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',moments=[1,2],
outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive',
mask='IRAS16293_Band9.H13CN_8_7.mask.subim',
chans='10~44',includepix=[0.4,100])
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative*')
immoments(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',moments=[1,2],
outfile='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative',
mask='IRAS16293_Band9.H13CN_8_7.mask.subim',
chans='10~44',includepix=[-100,-0.4])
Use CARTA to visualize the maps.
And finally export the result to FITS format for further analysis:
# In CASA
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.fits')
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.mom0.fits')
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_coord.fits')
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_coord.fits')
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_dispersion_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_dispersion_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Negative.weighted_dispersion_coord.fits')
os.system('rm -rf IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_dispersion_coord.fits')
exportfits(imagename='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_dispersion_coord',
fitsimage='IRAS16293_Band9.H13CN_8_7.image.subim.pbcor.Positive.weighted_dispersion_coord.fits')
Clean each spw
Some notes for this section: the threshold was chosen after doing a couple of smaller test images. This is about 3 sigma in channels with weak line emission. The mask was also chosen from a few test images. By setting the mask like this, there is no need to do interactive cleaning. Note however, that this clean mask does not do a good job on the extended emission of CO J=6-5. You may wish to experiment with the automasking algorithm in tclean.
You will see a warning that no rest frequency has been specified. You may wish to modify the rest frequency for each spw in order to have more accurate velocity labeling. A good reference for finding rest frequencies is splatalogue. Another good reference for Band 9 specifically is the 450 micron CSO survey of Orion: http://iopscience.iop.org/0067-0049/132/2/281/
If you would like to look at the initial cube and where the masks are located, use the tclean call in the loop to create an interactive clean session for one spw by setting 'interactive=True' and replacing 'str(i)' with the spw number you wish to clean. This will pop up an initial tclean window as before. Click through the cube channels using the arrow buttons in the 'Animator' section of the window. You can do one or more rounds of interactive cleaning, and then click the blue arrow under 'Next action' to let tclean finish non-interactively. Note that every time tclean needs to create a new cube for interactive viewing, it will take a while!
We first clean the first half of each spw.
# In CASA
os.system('rm -rf *spw*a.*')
spw=[0,1,2,3]
for i in spw:
tclean(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub',
imagename='IRAS16293_Band9.spw'+str(i)+'a',
spw=str(i),field='',phasecenter=3,
specmode='cube',outframe='LSRK',
nchan=950,start=20,width=2,
gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=False,restoringbeam='common',
mask=['circle[[16h32m22.61s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.86s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggsbwtaper',robust=0.5,pbcor=True,
niter=500, threshold='220.0mJy')
Next clean the 2nd half of each spw.
# In CASA
os.system('rm -rf *spw*b.*')
spw=[0,1,2,3]
for i in spw:
tclean(vis='IRAS16293_Band9.calibrated.tavg.ms.contsub',
imagename='IRAS16293_Band9.spw'+str(i)+'b',
spw=str(i),field='',phasecenter=3,
specmode='cube',outframe='LSRK',
nchan=950,start=1918,width=2,
gridder='mosaic',mosweight=True,
imsize=720,cell='0.028arcsec',
interactive=False,restoringbeam='common',
mask=['circle[[16h32m22.61s,-24d28m32.49s ], 0.75arcsec]','circle[[16h32m22.86s,-24d28m36.57s ], 1.0arcsec]'],
weighting='briggsbwtaper',robust=0.5,pbcor=True,
niter=500, threshold='220.0mJy')
Create subset images of the line cubes to center on the sources and export to FITS:
# In CASA
spw=[0,1,2,3]
for i in spw:
immath('IRAS16293_Band9.spw'+str(i)+'a.image.pbcor',
outfile='IRAS16293_Band9.spw'+str(i)+'a.image.subim.pbcor',
box='221,160,493,430',expr='IM0')
spw=[0,1,2,3]
for i in spw:
immath('IRAS16293_Band9.spw'+str(i)+'b.image.pbcor',
outfile='IRAS16293_Band9.spw'+str(i)+'b.image.subim.pbcor',
box='221,160,493,430',expr='IM0')
spw=[0,1,2,3]
for i in spw:
exportfits(imagename='IRAS16293_Band9.spw'+str(i)+'a.image.subim.pbcor',
fitsimage='IRAS16293_Band9.spw'+str(i)+'a.image.subim.pbcor.fits')
spw=[0,1,2,3]
for i in spw:
exportfits(imagename='IRAS16293_Band9.spw'+str(i)+'b.image.subim.pbcor',
fitsimage='IRAS16293_Band9.spw'+str(i)+'b.image.subim.pbcor.fits')