VLA Data Combination-W49A-CASA6.4.1: Difference between revisions
No edit summary |
mNo edit summary |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
''This | ''This CASA Guide is for Version 6.4.1 of CASA, and was last checked with CASA 6.5.4.'' | ||
== Introduction == | == Introduction == | ||
| Line 20: | Line 20: | ||
== The Data == | == The Data == | ||
We will be combining four different datasets, X-band A, B, C, and D configuration data. The data were all calibrated and the science target [https://casadocs.readthedocs.io/en/v6.4 | We will be combining four different datasets, X-band A, B, C, and D configuration data. The data were all calibrated and the science target [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.split.html#casatasks.manipulation.split split] out. | ||
The calibrated measurement sets can be downloaded here: | The calibrated measurement sets can be downloaded here: | ||
| Line 138: | Line 138: | ||
</pre> | </pre> | ||
The data were prepared for this tutorial to contain only one source, W49A, calibrated through the VLA pipeline (although, for the sake of this tutorial, no [https://casadocs.readthedocs.io/en/v6.4 | The data were prepared for this tutorial to contain only one source, W49A, calibrated through the VLA pipeline (although, for the sake of this tutorial, no [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] task has been run, see below). To reduce the size of the files, the MS only contains one spectral window, binned into 64 2MHz channels around 8.4GHz (X-band). We also extracted the calibrated data, discarding the raw data and any model, such that the MS now only contains visibilities in the DATA column. The on-source integration time amounts to about 2.5h. Inspection of the other array configuration files show almost identical setups. Although the integration times between the different array configurations do not follow the 1:3:9:27 ratios that we discussed in the previous section, we can still combine the data without any problem. In the end, having a higher signal to noise ratio on the shorter baselines can only improve the overall combined image. | ||
To better understand the data, let's check the uv-coverage of each of the four datasets. For faster plotting, we only plot channel 32 near the center of each spectral window. All plots are on the same scale: | To better understand the data, let's check the uv-coverage of each of the four datasets. For faster plotting, we only plot channel 32 near the center of each spectral window. All plots are on the same scale: | ||
| Line 169: | Line 169: | ||
The next step is to determine the image quality, the synthesized beam, and the rms of each image. The images do not have to be perfectly deconvolved as we only would like to see how the combination will improve over the individual arrays. | The next step is to determine the image quality, the synthesized beam, and the rms of each image. The images do not have to be perfectly deconvolved as we only would like to see how the combination will improve over the individual arrays. | ||
To keep things simple, we will use common cell sizes of 0.05 arcsec, which samples well the A-configuration beam and oversamples the more compact configurations. We create images of 1280 pixels on each side, and we will [https://casadocs.readthedocs.io/en/v6.4 | To keep things simple, we will use common cell sizes of 0.05 arcsec, which samples well the A-configuration beam and oversamples the more compact configurations. We create images of 1280 pixels on each side, and we will [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.imaging.tclean.html#casatasks.imaging.tclean tclean] 1000 iterations (see the [http://casaguides.nrao.edu/index.php/VLA_CASA_Imaging VLA CASA imaging guide] for more sophisticated imaging parameter choices). We move the center of the image to the region with the main emission to avoid having to image a very large field of view: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 193: | Line 193: | ||
The clean images, shown in Fig. | The clean images from each configuration, shown in Fig. 2, give a first impression of the data. Note that when combining the images, we will improve on the uv-coverage and will therefore not only combine high resolution with high surface brightness sensitivity on large scales, but also expect a higher image fidelity, i.e. fewer artifacts due to better deconvolution. In particular, the ripples in the extended configuration data are the typical 'missing short spacing' bowls, that the more compact configurations will fill in to improve the image quality. | ||
To display these images the CARTA software was used. We would like to note that the CASAviewer has not been maintained for a few years anymore and will be removed from future versions of CASA. | |||
The NRAO replacement visualization tool for images and cubes is CARTA, the “Cube Analysis and Rendering Tool for Astronomy”. It is available from the CARTA website [https://cartavis.org/ carta]. We strongly recommend to use CARTA, as it provides a much more efficient, stable, and feature rich user experience. A comparison of the CASAviewer and CARTA, as well as instructions on how to use CARTA at NRAO is provided in the respective section of CASA docs https://casadocs.readthedocs.io/en/stable/notebooks/carta.html. This tutorial shows Figures generated with CARTA for visualization. | |||
{| | {| | ||
|[[Image: | |[[Image:W49A_Aonly_Bonly_Conly_Donly_CASA6.4.1.png|1400px|thumb|left|'''Figure 2:''' <br />W49A in each VLA configuration.]] | ||
|} | |} | ||
The basic parameters of the individual images can be checked through [https://casadocs.readthedocs.io/en/v6.4 | The basic parameters of the individual images can be checked through [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.information.imhead.html#casatasks.information.imhead imhead] and the beam sizes are 0.19"x0.19" for A, 0.71"x0.66" for B, 2.20"x2.04" for C and 8.44"x6.36" for D-configuration. This reflects again the factor of three in resolution between the different arrays. | ||
== Check and Adjust the Visibility Weights == | == Check and Adjust the Visibility Weights == | ||
| Line 248: | Line 248: | ||
1) Calculate the weights based on the rms of the visibilities themselves, using the [https://casadocs.readthedocs.io/en/v6.4 | 1) Calculate the weights based on the rms of the visibilities themselves, using the [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] task. | ||
2) Reset the weights with [https://casadocs.readthedocs.io/en/v6.4 | 2) Reset the weights with [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.calibration.initweights.html#casatasks.calibration.initweights initweights] to reflect the channel width and correlator integration time (<math>WEIGHT=2 \Delta \nu \Delta t</math>, see the document on [https://casadocs.readthedocs.io/en/stable/notebooks/data_weights.html Data Weights] in CASAdocs). | ||
3) Calculate the weights based on the rms of the cross-polarization products (currently not supported in [https://casadocs.readthedocs.io/en/v6.4 | 3) Calculate the weights based on the rms of the cross-polarization products (currently not supported in [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt]). | ||
We recommend to reset the weights when there are strong sources present in the data as they will change the rms of the visibilities and the rms will not be representative of the noise anymore. Without strong sources, [https://casadocs.readthedocs.io/en/v6.4 | We recommend to reset the weights when there are strong sources present in the data as they will change the rms of the visibilities and the rms will not be representative of the noise anymore. Without strong sources, [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] should deliver better results. The third option may work for both cases, but requires full polarization observations and calibrations. For this guide we will follow the [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] path as an example. But the user should be aware of the different options for optimized imaging. | ||
| Line 260: | Line 260: | ||
[https://casadocs.readthedocs.io/en/v6.4 | [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] will bring all the weights of all observations on the same scale. The task recalculates the visibility weights based on the inverse of their rms. Task [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] is part of the [https://go.nrao.edu/vla-pipe VLA pipeline], so the pipelined data may already have recalculated weights and this step can be skipped. It does not hurt though, to re-run [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt]. | ||
For more information on weights, see [https://casadocs.readthedocs.io/en/stable/notebooks/data_weights.html definition of data weights]. | For more information on weights, see [https://casadocs.readthedocs.io/en/stable/notebooks/data_weights.html definition of data weights]. | ||
Task [https://casadocs.readthedocs.io/en/v6.4 | Task [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] will be executed on each MS. The default setting calculates the weight based on the rms of each scan and spectral window. This setting works quite well for continuum observations. We would like to note though that for spectral line data the ''fitspw'' parameter should be set to exclude the line from the calculations. Otherwise, strong lines will be down-weighted. | ||
Now, we execute [https://casadocs.readthedocs.io/en/v6.4 | Now, we execute [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt]. Since the DATA column is our calibrated data (see above; in a general case it would be the CORRECTED_DATA column), we calculate the rms based on the values in that column: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 282: | Line 282: | ||
</source> | </source> | ||
Let's have a look at the weight values again to see if [https://casadocs.readthedocs.io/en/v6.4 | Let's have a look at the weight values again to see if [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.statwt.html#casatasks.manipulation.statwt statwt] has adjusted the weights properly: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 313: | Line 313: | ||
We will now create a combined image of all four re-weighted datasets. | We will now create a combined image of all four re-weighted datasets. | ||
First, let's check the new uv-coverage. We concatenate the data with [https://casadocs.readthedocs.io/en/v6.4 | First, let's check the new uv-coverage. We concatenate the data with [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.concat.html#casatasks.manipulation.concat concat]: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 334: | Line 334: | ||
Although [https://casadocs.readthedocs.io/en/v6.4 | Although [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.concat.html#casatasks.manipulation.concat concat] merges all four MSs into a single one, it is actually not a required step before imaging. Task [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.imaging.tclean.html#casatasks.imaging.tclean tclean] will take care of the combination when all datasets are specified as a list. Tclean will also perform the spectral regridding of all datasets on the fly, in particular in ''mode="velocity"'' or ''mode="frequency"''. There's no need to run [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.cvel.html#casatasks.manipulation.cvel cvel]/[https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.cvel2.html#casatasks.manipulation.cvel2 cvel2] (or [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.mstransform.html#casatasks.manipulation.mstransform mstransform]) to Doppler correct the MSs beforehand. | ||
We will now create a combined image in [https://casadocs.readthedocs.io/en/v6.4 | We will now create a combined image in [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.imaging.tclean.html#casatasks.imaging.tclean tclean]. The ''threshold'' parameter was derived by a previous run of [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.imaging.tclean.html#casatasks.imaging.tclean tclean] on the combined MS for which we determined the rough rms noise. For our threshold, we will use the rms noise multiplied by a factor of ~2.5. | ||
| Line 347: | Line 347: | ||
</source> | </source> | ||
To display this image the CARTA software was used. | |||
{| | {| | ||
|[[Image: | |[[Image:W49A_combinedABCD_CASA6.4.1.png|400px|thumb|left|'''Figure 6:''' <br />Combined image. ]] | ||
|} | |} | ||
The combined beam is now 0.26"x0.24". | The combined beam is now 0.26"x0.24". | ||
The image (Fig. 6) can still be improved upon. For simplicity, we did not use any interactive cleaning in the above, but we highly recommend it for producing the final images. Improvements can also be obtained by adjusting the image weights via the Briggs robust parameter, adding a taper, or weighting the different datasets against each other using ''visweightscale'' in [https://casadocs.readthedocs.io/en/v6.4 | The image (Fig. 6) can still be improved upon. For simplicity, we did not use any interactive cleaning in the above, but we highly recommend it for producing the final images. Improvements can also be obtained by adjusting the image weights via the Briggs robust parameter, adding a taper, or weighting the different datasets against each other using ''visweightscale'' in [https://casadocs.readthedocs.io/en/v6.5.4/api/tt/casatasks.manipulation.concat.html#casatasks.manipulation.concat concat]. Wide-band imaging and multi-scale imaging will also lead to better results. We refer to the [http://casaguides.nrao.edu/index.php/VLA_CASA_Imaging VLA CASA Imaging Guide] for more details and examples. | ||
== Tips for Selfcal == | == Tips for Selfcal == | ||
If the source has a bright nucleus or, more generally, a bright unresolved emission, start with the A array data, selfcal, then add B array, selfcal again, and so on. This procedure starts with a high model flux that is increased further. If the source is mostly diffuse, then there is not much signal in the A array data, so start with the D array, selfcal, then add C array, selfcal, and so on. Each selfcal step should be phase-only first with maybe two or more iterations. At the end of each selfcal step, a phase+amplitude selfcal can be attempted, before merging in the next array configuration data. After each merge the selfcal steps can be repeated. General selfcal procedures are outlined in the [https://casaguides.nrao.edu/index.php?title=Karl_G._Jansky_VLA_Tutorials#Self-calibration_of_VLA_Data Self-Calibration topical guide]. | If the source has a bright nucleus or, more generally, a bright unresolved emission, start with the A array data, selfcal, then add B array, selfcal again, and so on. This procedure starts with a high model flux that is increased further. If the source is mostly diffuse, then there is not much signal in the A array data, so start with the D array, selfcal, then add C array, selfcal, and so on. Each selfcal step should be phase-only first with maybe two or more iterations. At the end of each selfcal step, a phase+amplitude selfcal can be attempted, before merging in the next array configuration data. After each merge the selfcal steps can be repeated. General selfcal procedures are outlined in the [https://casaguides.nrao.edu/index.php?title=Karl_G._Jansky_VLA_Tutorials#Self-calibration_of_VLA_Data Self-Calibration topical guide]. | ||
<!-- | <!-- | ||
| Line 367: | Line 365: | ||
Edited: Updated to CASA 5.5.0, Tony Perreault May 2019 | Edited: Updated to CASA 5.5.0, Tony Perreault May 2019 | ||
Changed into W49A tutorial: Juergen Ott September 2021 | Changed into W49A tutorial: Juergen Ott September 2021 | ||
Edited: Updated to CASA 6.5.4, checked in 6.5.4, James Khor January 2024 | |||
--> | --> | ||
{{Checked 6.4 | == Extracted Scripts for VLA Data Combination-W49A-CASA6.5.4 == | ||
[[Media:VLADataCombination-W49A-CASA6.4.1.txt|VLADataCombination-W49A-CASA6.5.4.txt]] | |||
{{Checked 6.5.4}} | |||
Latest revision as of 20:55, 22 February 2024
This CASA Guide is for Version 6.4.1 of CASA, and was last checked with CASA 6.5.4.
Introduction
A perfect image requires measurement of all spatial scales, which is equivalent to filling in the complete uv plane. Unfortunately, this can never be achieved with an aperture synthesis interferometer, although a large number of baselines, long integration times, and multi-frequency-synthesis are all good approaches to increase the uv-coverage. One method to obtain more baselines is to observe in different array configurations and to combine the data afterwards. Deconvolution algorithms are then given good starting conditions to interpolate across the gaps in the uv-plane to achieve an image that combines the surface brightness sensitivity of the compact baselines with the spatial resolution of the extended baselines.
The VLA can be configured into four principal array configurations: A, B, C, and D. The A configuration is the most extended, and D is the most compact configuration. Consequently, A-configuration data exhibit the highest spatial resolution whereas D-configuration delivers the best surface brightness sensitivity and also images the largest scales of the sky brightness distribution.
In this tutorial, we will combine data from all four VLA configurations (A, B, C, and D) that were observed in X-band continuum toward W49A, a massive star forming region in the Milky Way.
Typical Observing Times
When an object is being observed by the VLA in different array configurations, on-source integration times are ideally matched to reach a common surface brightness sensitivity for all scales. Adjacent VLA configurations result in synthesized beams that differ in linear size by approximately a factor of 3. The beam areas therefore change by factors of ~10 and the more extended configuration would ideally need to have 10 times more integration time than the next compact one. This, however, is frequently not very practical and it turns out that integration times that differ by factors of ~3 deliver data that can be satisfactorily combined. This combination matches the sensitivity of overlapping VLA visibilities when the data are convolved to the same scales.
Although overlapping uv-coverages are essential for the best imaging, it is possible to combine non-overlapping data if one understands that some spacings are not present and that the adjustment of the individual datasets is somewhat subjective. Weighting will be primarily achieved during imaging by the Briggs scheme that allows one to adjust imaging weights between the natural (weighting by the number of visibilities that are gridded in each cell) and uniform (weighting by the cells themselves) extremes.
Additionally, each visibility exhibits weights that should only depend on correlator integration time, bandwidth, and system temperature (Tsys). Note that the VLA does not apply Tsys in the online system. Visibility weights between different observations will therefore need to be adjusted, as described below, before they can be combined.
The Data
We will be combining four different datasets, X-band A, B, C, and D configuration data. The data were all calibrated and the science target split out.
The calibrated measurement sets can be downloaded here: https://casa.nrao.edu/Data/EVLA/combination/VLA-combination-W49A.tar.gz (3.0G)
As a first step download the file above, then untar:
# In a Terminal tar -xzvf VLA-combination-W49A.tar.gz
This will unpack four MeasurementSets (MSs), one for each array configuration:
A-W49A.ms B-W49A.ms C-W49A.ms D-W49A.ms
Initial Imaging
We will inspect the data and create separate images to better understand the image parameters such as on-source integration time, resolution, and rms.
Start CASA:
# In a Terminal casa
As a first step, let's have a look at the 'listobs' output for the different MSs. The listobs task will output a dictionary to the terminal and a table to the log. For example, the A-configuration MS has the following structure:
# In CASA
listobs(vis='A-W49A.ms')
##########################################
##### Begin Task: listobs #####
listobs( vis='A-W49A.ms', selectdata=True, spw='', field='', antenna='', uvrange='', timerange='', correlation='', scan='', intent='', feed='', array='', observation='', verbose=True, listfile='', listunfl=False, cachesize=50.0, overwrite=False )
================================================================================
MeasurementSet Name: /lustre/aoc/sciops/jott/casa/combination-2021/data/vis-small3/A-W49A.ms MS Version 2
================================================================================
Observer: Dr. Chris De Pree Project: uid://evla/pdb/30105074
Observation: EVLA
Computing scan and subscan properties...
Data records: 764478 Total elapsed time = 8973 seconds
Observed from 24-Jun-2015/07:32:30.0 to 24-Jun-2015/10:02:03.0 (UTC)
ObservationID = 0 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent
24-Jun-2015/07:32:30.0 - 07:42:24.0 4 0 W49A 69498 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
07:42:27.0 - 07:47:24.0 5 0 W49A 34749 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
07:49:27.0 - 07:59:21.0 7 0 W49A 69498 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
07:59:24.0 - 08:04:21.0 8 0 W49A 34749 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:06:24.0 - 08:16:18.0 10 0 W49A 69498 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:16:21.0 - 08:21:18.0 11 0 W49A 34749 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:23:21.0 - 08:33:18.0 13 0 W49A 69849 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:33:21.0 - 08:38:15.0 14 0 W49A 34398 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:40:18.0 - 08:50:15.0 16 0 W49A 69849 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:50:18.0 - 08:55:12.0 17 0 W49A 34398 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
08:57:15.0 - 09:07:12.0 19 0 W49A 69849 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
09:07:15.0 - 09:12:09.0 20 0 W49A 34398 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
09:40:09.0 - 09:50:03.0 25 0 W49A 69498 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
09:50:06.0 - 09:55:03.0 26 0 W49A 34749 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
09:57:06.0 - 10:02:03.0 28 0 W49A 34749 [0] [3] [OBSERVE_TARGET#UNSPECIFIED]
(nRows = Total number of rows per scan)
Fields: 1
ID Code Name RA Decl Epoch SrcId nRows
0 NONE W49A 19:10:12.930999 +09.06.11.88200 J2000 0 764478
Spectral Windows: (1 unique spectral windows and 1 unique polarization setups)
SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) BBC Num Corrs
0 EVLA_X#B0D0#11 64 TOPO 8372.479 2000.000 128000.0 8435.4793 15 RR LL
Sources: 1
ID Name SpwId RestFreq(MHz) SysVel(km/s)
0 W49A 0 9816.865 8
Antennas: 27:
ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m)
East North Elevation x y z
0 ea01 N64 25.0 m -107.37.58.7 +34.02.20.5 -1382.3871 15410.1384 -40.6410 -1599855.687000 -5033332.368600 3567636.613800
1 ea02 N16 25.0 m -107.37.10.9 +33.54.48.0 -155.8524 1426.6393 -9.3842 -1601061.957600 -5041175.881400 3556058.033100
2 ea03 W48 25.0 m -107.42.44.3 +33.50.52.1 -8707.9476 -5861.7878 15.5283 -1610451.932800 -5042471.123800 3550021.055800
3 ea04 E40 25.0 m -107.32.35.4 +33.52.16.9 6908.8199 -3240.7429 39.0197 -1595124.937100 -5045829.476200 3552210.683600
4 ea05 E72 25.0 m -107.24.42.3 +33.49.18.0 19041.8648 -8769.1806 4.7639 -1584460.883200 -5052385.614800 3547600.041100
5 ea06 N40 25.0 m -107.37.29.5 +33.57.44.4 -633.6056 6878.6057 -20.7877 -1600592.749000 -5038121.341300 3560574.846200
6 ea07 N72 25.0 m -107.38.10.5 +34.04.12.2 -1685.6797 18861.8360 -43.4978 -1599557.928700 -5031396.353400 3570494.743400
7 ea08 E64 25.0 m -107.27.00.1 +33.50.06.7 15507.6019 -7263.7123 67.2037 -1587600.192200 -5050575.870700 3548885.413900
8 ea09 W08 25.0 m -107.37.21.6 +33.53.53.0 -432.1156 -272.1458 -1.4994 -1601614.091200 -5042001.656900 3554652.514300
9 ea10 E32 25.0 m -107.34.01.5 +33.52.50.3 4701.6612 -2209.7048 25.2200 -1597053.118400 -5044604.692200 3553059.011100
10 ea11 W24 25.0 m -107.38.49.0 +33.53.04.0 -2673.3543 -1784.5861 10.4723 -1604008.747100 -5042135.805100 3553403.716300
11 ea12 N32 25.0 m -107.37.22.0 +33.56.33.6 -441.7251 4689.9820 -16.9255 -1600781.044100 -5039347.437000 3558761.543300
12 ea13 W16 25.0 m -107.37.57.4 +33.53.33.0 -1348.7131 -890.6107 1.2990 -1602592.853500 -5042054.989100 3554140.713000
13 ea14 E08 25.0 m -107.36.48.9 +33.53.55.1 407.8280 -206.0296 -3.2233 -1600801.931400 -5042219.381700 3554706.431200
14 ea15 E24 25.0 m -107.35.13.4 +33.53.18.1 2858.1759 -1349.1337 13.7125 -1598663.094300 -5043581.381100 3553767.012000
15 ea16 W64 25.0 m -107.46.20.1 +33.48.50.9 -14240.7524 -9606.2900 17.0885 -1616361.575500 -5042770.516600 3546911.419900
16 ea17 N24 25.0 m -107.37.16.1 +33.55.37.7 -290.3645 2961.8847 -12.2406 -1600930.072900 -5040316.384900 3557330.407200
17 ea18 W72 25.0 m -107.48.24.0 +33.47.41.2 -17419.4641 -11760.2694 14.9442 -1619757.299900 -5042937.656400 3545120.392300
18 ea19 W40 25.0 m -107.41.13.5 +33.51.43.1 -6377.9723 -4286.7839 8.2107 -1607962.451800 -5042338.204100 3551324.945500
19 ea20 N48 25.0 m -107.37.38.1 +33.59.06.2 -855.2671 9405.9613 -25.9164 -1600374.881000 -5036704.217500 3562667.893900
20 ea21 E56 25.0 m -107.29.04.1 +33.50.54.9 12327.6481 -5774.7445 42.6332 -1590380.599000 -5048810.261300 3550108.444300
21 ea23 E16 25.0 m -107.36.09.8 +33.53.40.0 1410.0345 -673.4704 -0.7961 -1599926.106100 -5042772.964400 3554319.787600
22 ea24 W32 25.0 m -107.39.54.8 +33.52.27.2 -4359.4410 -2923.1315 11.7693 -1605808.637100
23 ea25 W56 25.0 m -107.44.26.7 +33.49.54.6 -11333.1991 -7637.6832 15.3636 -1613255.391400 -5042613.097800 3548545.906000
24 ea26 E48 25.0 m -107.30.56.1 +33.51.38.4 9456.6036 -4431.6334 37.9266 -1592894.077600 -5047229.118200 3551221.221000
25 ea27 N08 25.0 m -107.37.07.5 +33.54.15.8 -68.9101 433.1984 -5.0732 -1601147.939700 -5041733.820400 3555235.956600
26 ea28 N56 25.0 m -107.37.47.9 +34.00.38.4 -1105.2275 12254.3062 -34.2445 -1600128.402500 -5035104.139200 3565024.670400
Result listobs: True
Task listobs complete. Start time: 2021-09-10 10:01:51.648534 End time: 2021-09-10 10:01:51.932094
##### End Task: listobs #####
##########################################
The data were prepared for this tutorial to contain only one source, W49A, calibrated through the VLA pipeline (although, for the sake of this tutorial, no statwt task has been run, see below). To reduce the size of the files, the MS only contains one spectral window, binned into 64 2MHz channels around 8.4GHz (X-band). We also extracted the calibrated data, discarding the raw data and any model, such that the MS now only contains visibilities in the DATA column. The on-source integration time amounts to about 2.5h. Inspection of the other array configuration files show almost identical setups. Although the integration times between the different array configurations do not follow the 1:3:9:27 ratios that we discussed in the previous section, we can still combine the data without any problem. In the end, having a higher signal to noise ratio on the shorter baselines can only improve the overall combined image.
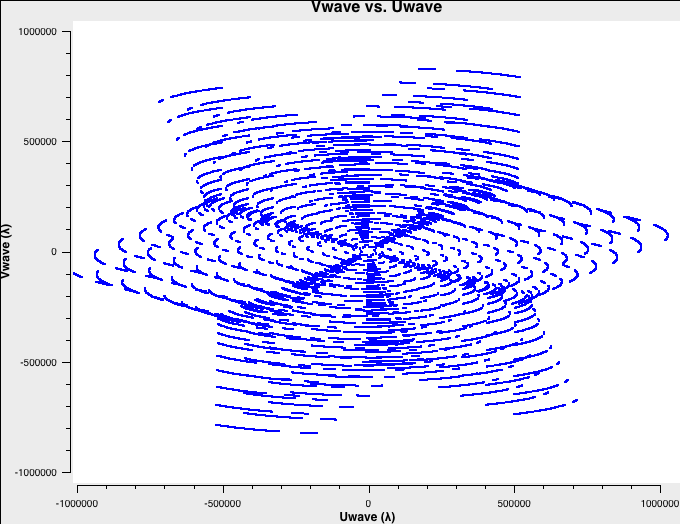
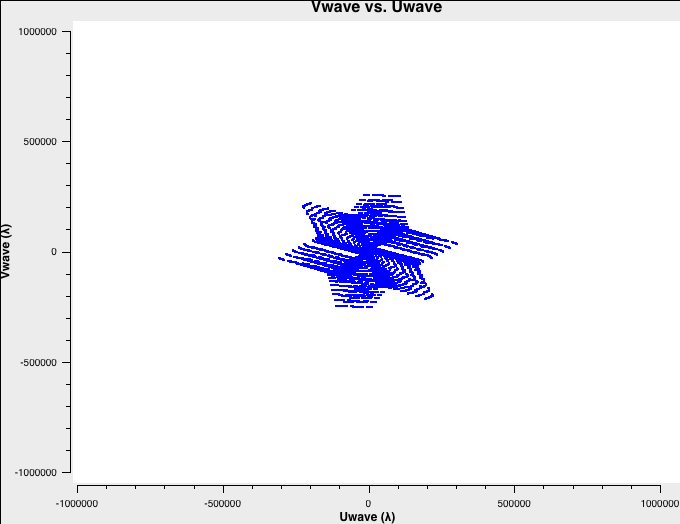
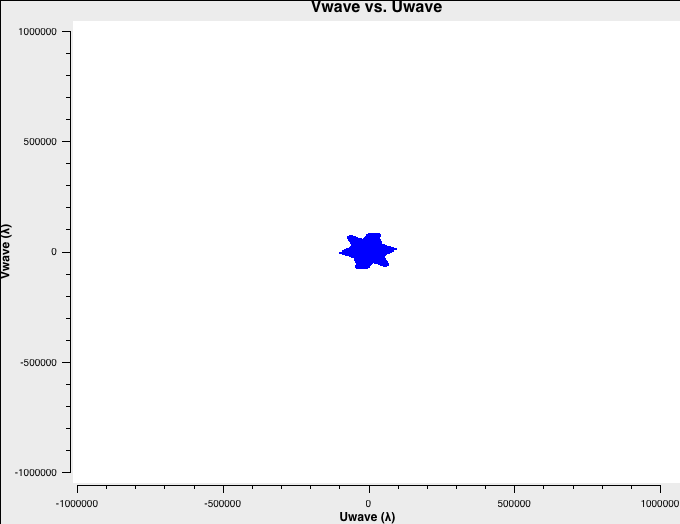
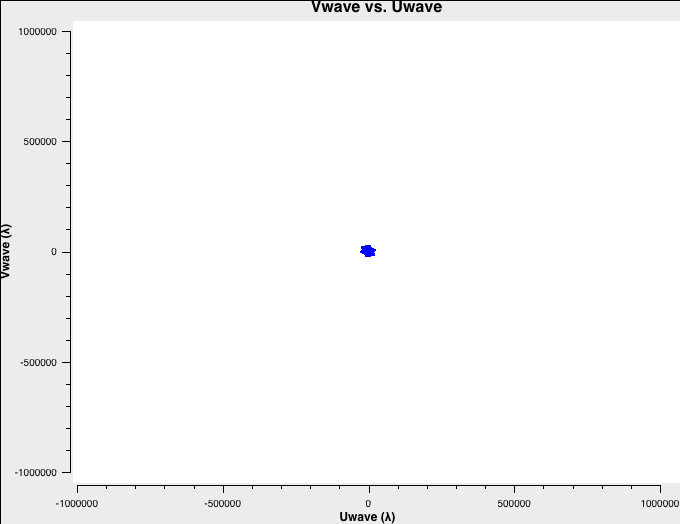
To better understand the data, let's check the uv-coverage of each of the four datasets. For faster plotting, we only plot channel 32 near the center of each spectral window. All plots are on the same scale:
# In CASA
# A-config:
plotms(vis='A-W49A.ms',xaxis='Uwave',yaxis='Vwave',spw='0:32',plotrange=[-1000000,1000000,-1000000,1000000])
# B-config
plotms(vis='B-W49A.ms',xaxis='Uwave',yaxis='Vwave',spw='0:32',plotrange=[-1000000,1000000,-1000000,1000000])
# C-config:
plotms(vis='C-W49A.ms',xaxis='Uwave',yaxis='Vwave',spw='0:32',plotrange=[-1000000,1000000,-1000000,1000000])
# D-config:
plotms(vis='D-W49A.ms',xaxis='Uwave',yaxis='Vwave',spw='0:32',plotrange=[-1000000,1000000,-1000000,1000000])
The uv-coverage plots are shown in Fig. 1a-d using a common scale. The longest baseline in each array differs by about a factor of 3, as expected, between the VLA A, B, C, and D configurations.
The next step is to determine the image quality, the synthesized beam, and the rms of each image. The images do not have to be perfectly deconvolved as we only would like to see how the combination will improve over the individual arrays.
To keep things simple, we will use common cell sizes of 0.05 arcsec, which samples well the A-configuration beam and oversamples the more compact configurations. We create images of 1280 pixels on each side, and we will tclean 1000 iterations (see the VLA CASA imaging guide for more sophisticated imaging parameter choices). We move the center of the image to the region with the main emission to avoid having to image a very large field of view:
# In CASA
# A-config:
tclean(vis='A-W49A.ms',imagename='Aonly',cell='0.05arcsec',imsize=[1280,1280],weighting='briggs',
robust=0,specmode='mfs',phasecenter='J2000 19:10:14 +09.06.13.7')
# B-config:
tclean(vis='B-W49A.ms',imagename='Bonly',cell='0.05arcsec',imsize=[1280,1280],weighting='briggs',
robust=0,specmode='mfs',phasecenter='J2000 19:10:14 +09.06.13.7')
# C-config:
tclean(vis='C-W49A.ms',imagename='Conly',cell='0.05arcsec',imsize=[1280,1280],weighting='briggs',
robust=0,specmode='mfs',phasecenter='J2000 19:10:14 +09.06.13.7')
# D-config:
tclean(vis='D-W49A.ms',imagename='Donly',cell='0.05arcsec',imsize=[1280,1280],weighting='briggs',
robust=0,specmode='mfs',phasecenter='J2000 19:10:14 +09.06.13.7')
The clean images from each configuration, shown in Fig. 2, give a first impression of the data. Note that when combining the images, we will improve on the uv-coverage and will therefore not only combine high resolution with high surface brightness sensitivity on large scales, but also expect a higher image fidelity, i.e. fewer artifacts due to better deconvolution. In particular, the ripples in the extended configuration data are the typical 'missing short spacing' bowls, that the more compact configurations will fill in to improve the image quality.
To display these images the CARTA software was used. We would like to note that the CASAviewer has not been maintained for a few years anymore and will be removed from future versions of CASA. The NRAO replacement visualization tool for images and cubes is CARTA, the “Cube Analysis and Rendering Tool for Astronomy”. It is available from the CARTA website carta. We strongly recommend to use CARTA, as it provides a much more efficient, stable, and feature rich user experience. A comparison of the CASAviewer and CARTA, as well as instructions on how to use CARTA at NRAO is provided in the respective section of CASA docs https://casadocs.readthedocs.io/en/stable/notebooks/carta.html. This tutorial shows Figures generated with CARTA for visualization.
 W49A in each VLA configuration. |
The basic parameters of the individual images can be checked through imhead and the beam sizes are 0.19"x0.19" for A, 0.71"x0.66" for B, 2.20"x2.04" for C and 8.44"x6.36" for D-configuration. This reflects again the factor of three in resolution between the different arrays.
Check and Adjust the Visibility Weights
VLA visibility weights are currently based only on channel width and correlator integration time. In the future, the VLA may use the switched power measurements to derive absolute calibrated weights. At the moment, however, the VLA weights need to be taken as being relative to each other. The relative sensitivity within an observation is measured by the gain, so weights of single, continuous observations are self-consistent. It is important to adjust the weights between separate observations as they will potentially be on different scales.
Let's have a look at the weights of the different datasets. We will plot the weights as a function of uv-distance in a single central channel for improved plotting time:
# In CASA
# A-config:
plotms(vis='A-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
# In CASA
# B-config
plotms(vis='B-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
# In CASA
# C-config:
plotms(vis='C-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
# In CASA
# D-config:
plotms(vis='D-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
Fig. 3a-d shows the weights and they are as described above.
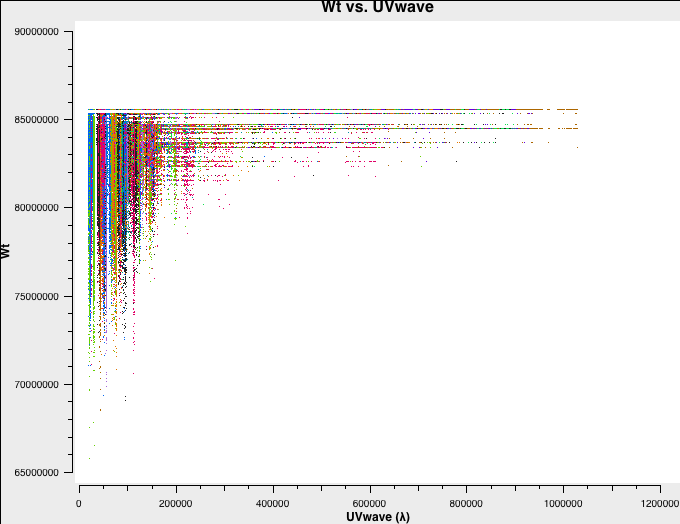 A-config original weights. |
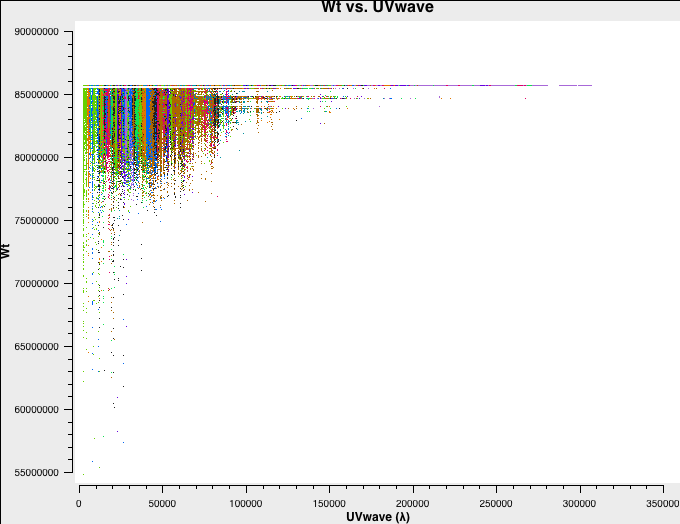 B-config original weights. |
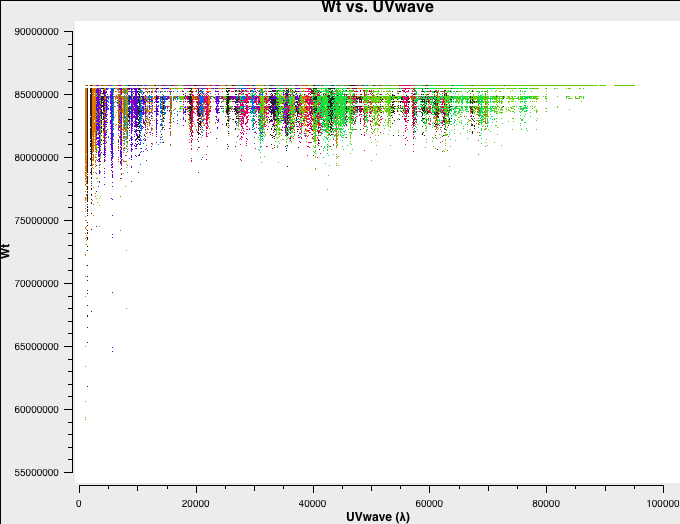 C-config original weights. |
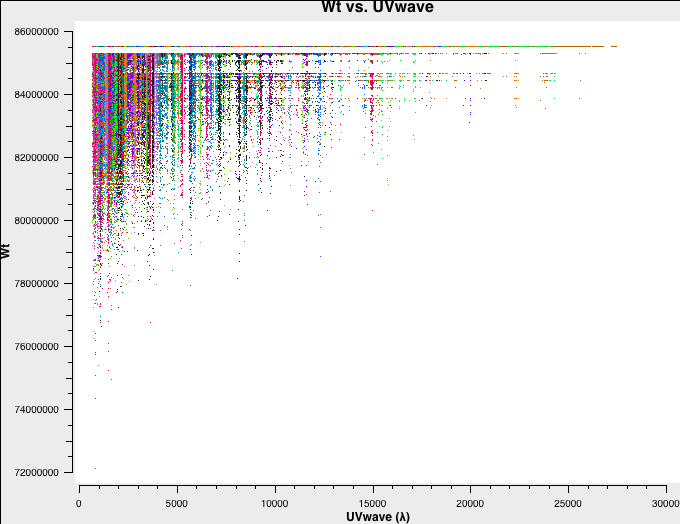 D-config original weights. |
The next step is to bring the weights on the same relative scale.
There are currently a number of options to do so.
1) Calculate the weights based on the rms of the visibilities themselves, using the statwt task.
2) Reset the weights with initweights to reflect the channel width and correlator integration time ([math]\displaystyle{ WEIGHT=2 \Delta \nu \Delta t }[/math], see the document on Data Weights in CASAdocs).
3) Calculate the weights based on the rms of the cross-polarization products (currently not supported in statwt).
We recommend to reset the weights when there are strong sources present in the data as they will change the rms of the visibilities and the rms will not be representative of the noise anymore. Without strong sources, statwt should deliver better results. The third option may work for both cases, but requires full polarization observations and calibrations. For this guide we will follow the statwt path as an example. But the user should be aware of the different options for optimized imaging.
Calculating the weights based on the rms
statwt will bring all the weights of all observations on the same scale. The task recalculates the visibility weights based on the inverse of their rms. Task statwt is part of the VLA pipeline, so the pipelined data may already have recalculated weights and this step can be skipped. It does not hurt though, to re-run statwt. For more information on weights, see definition of data weights.
Task statwt will be executed on each MS. The default setting calculates the weight based on the rms of each scan and spectral window. This setting works quite well for continuum observations. We would like to note though that for spectral line data the fitspw parameter should be set to exclude the line from the calculations. Otherwise, strong lines will be down-weighted.
Now, we execute statwt. Since the DATA column is our calibrated data (see above; in a general case it would be the CORRECTED_DATA column), we calculate the rms based on the values in that column:
# In CASA
# A-config:
statwt(vis='A-W49A.ms',datacolumn='data')
# B-config
statwt(vis='B-W49A.ms',datacolumn='data')
# C-config:
statwt(vis='C-W49A.ms',datacolumn='data')
# D-config:
statwt(vis='D-W49A.ms',datacolumn='data')
Let's have a look at the weight values again to see if statwt has adjusted the weights properly:
# In CASA
# A-config:
plotms(vis='A-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
# B-config
plotms(vis='B-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
# C-config:
plotms(vis='C-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
# D-config:
plotms(vis='D-W49A.ms',spw='0:32',xaxis='uvwave',yaxis='wt',coloraxis='antenna1')
In Fig. 4a-d we plot the new, recalculated weights for the three configurations. The absolute scaling has indeed changed. and is now in units of 1/Jy^2.
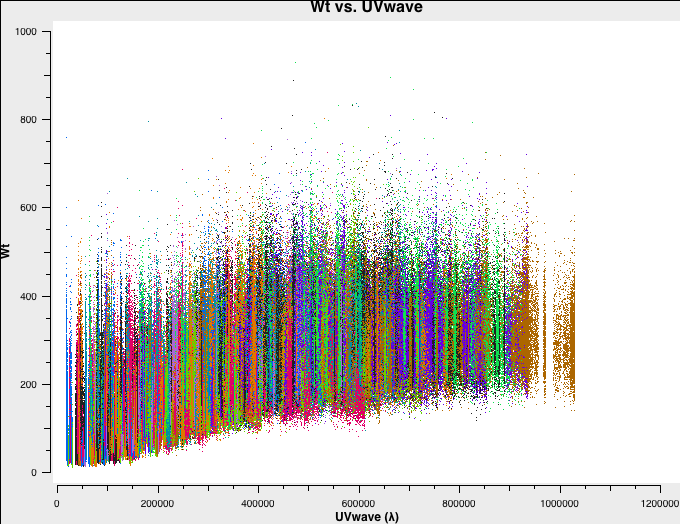 A-config recalculated weights. |
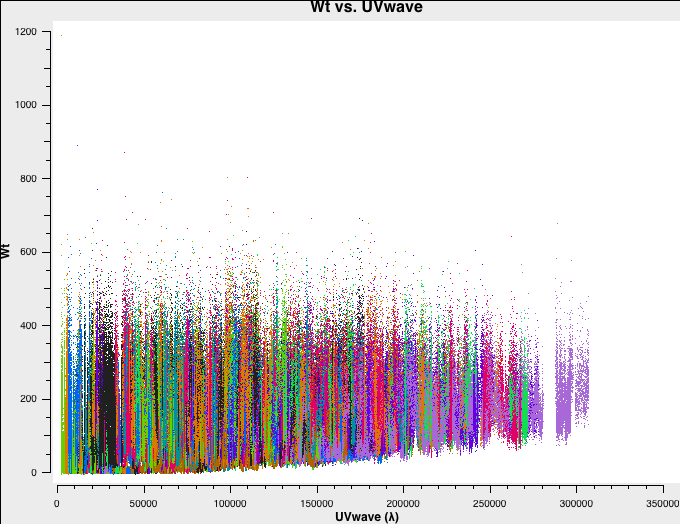 B-config recalculated weights. |
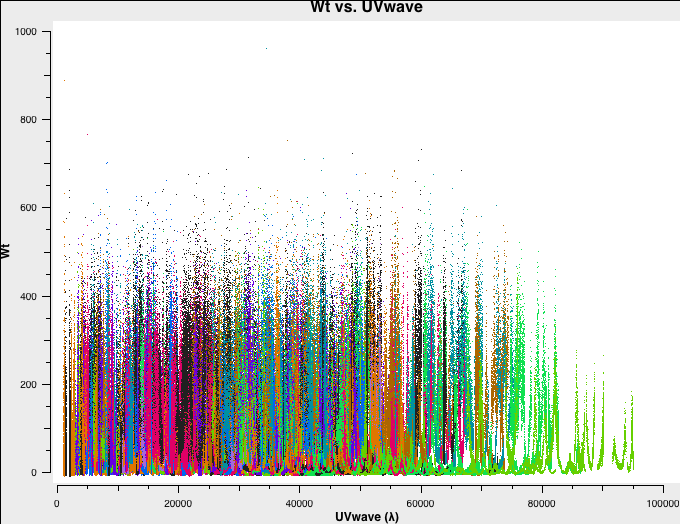 C-config recalculated weights. |
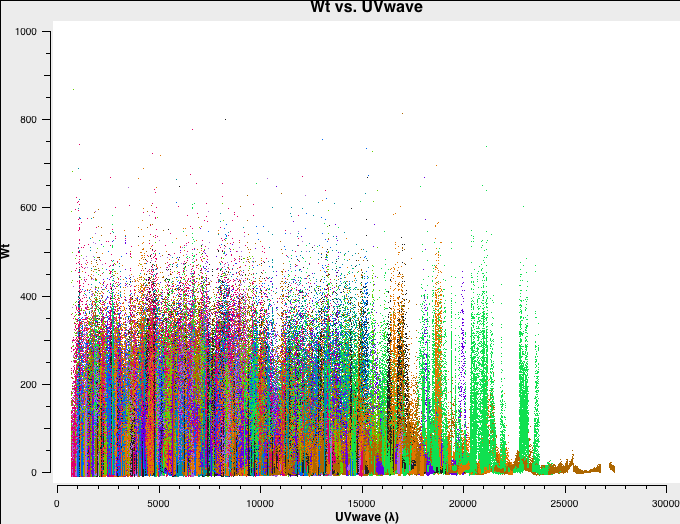 D-config recalculated weights. |
Combining and Imaging All Data
We will now create a combined image of all four re-weighted datasets.
First, let's check the new uv-coverage. We concatenate the data with concat:
# In CASA
concat(vis=['A-W49A.ms','B-W49A.ms','C-W49A.ms','D-W49A.ms'],concatvis='ABCD-W49A.ms')
Now let's plot the uv-coverage of the combined MSs. Since the spws have been renumbered, we plot the central channel of each sub-configuration with the spw='*:32' keyword:
# In CASA
plotms(vis='ABCD-W49A.ms',xaxis='Uwave',yaxis='Vwave',spw='*:32')
Fig. 5 shows the combined uv-coverage, which extends to A-configuration baselines but with a much higher density at the intermediate and short baselines contributed from B, C, and D configurations.
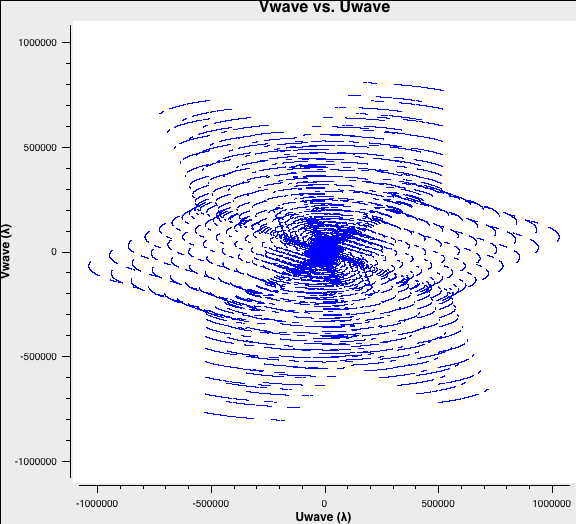 Combined uv-coverage. |
Although concat merges all four MSs into a single one, it is actually not a required step before imaging. Task tclean will take care of the combination when all datasets are specified as a list. Tclean will also perform the spectral regridding of all datasets on the fly, in particular in mode="velocity" or mode="frequency". There's no need to run cvel/cvel2 (or mstransform) to Doppler correct the MSs beforehand.
We will now create a combined image in tclean. The threshold parameter was derived by a previous run of tclean on the combined MS for which we determined the rough rms noise. For our threshold, we will use the rms noise multiplied by a factor of ~2.5.
Image the data with recalculated weights:
# In CASA
tclean(vis=['ABCD-W49A.ms'], imagename='W49A_combinedABCD',specmode='mfs',cell='0.05arcsec',imsize=[1280,1280],
niter=10000, weighting='briggs',robust=0, phasecenter='J2000 19:10:14 +09.06.13.7',threshold='2mJy')
To display this image the CARTA software was used.
 Combined image. |
The combined beam is now 0.26"x0.24". The image (Fig. 6) can still be improved upon. For simplicity, we did not use any interactive cleaning in the above, but we highly recommend it for producing the final images. Improvements can also be obtained by adjusting the image weights via the Briggs robust parameter, adding a taper, or weighting the different datasets against each other using visweightscale in concat. Wide-band imaging and multi-scale imaging will also lead to better results. We refer to the VLA CASA Imaging Guide for more details and examples.
Tips for Selfcal
If the source has a bright nucleus or, more generally, a bright unresolved emission, start with the A array data, selfcal, then add B array, selfcal again, and so on. This procedure starts with a high model flux that is increased further. If the source is mostly diffuse, then there is not much signal in the A array data, so start with the D array, selfcal, then add C array, selfcal, and so on. Each selfcal step should be phase-only first with maybe two or more iterations. At the end of each selfcal step, a phase+amplitude selfcal can be attempted, before merging in the next array configuration data. After each merge the selfcal steps can be repeated. General selfcal procedures are outlined in the Self-Calibration topical guide.
Extracted Scripts for VLA Data Combination-W49A-CASA6.5.4
VLADataCombination-W49A-CASA6.5.4.txt
Last checked on CASA Version 6.5.4