VLA CASA Flagging-CASA5.4.0: Difference between revisions
| (21 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
This CASA guide covers online data flagging, shadowing, zero-clipping, and quacking. It also covers auto-flagging RFI (Radio Frequency Interference) via TFCrop (Time-Frequency Crop) and rflag. | This CASA guide covers online data flagging, shadowing, zero-clipping, and quacking. It also covers auto-flagging RFI (Radio Frequency Interference) via TFCrop (Time-Frequency Crop) and rflag. | ||
We will be utilizing data taken, with the Karl G. Jansky Very Large Array, of [http://simbad.u-strasbg.fr/simbad/sim-id?Ident=SNR+G055.7%2B03.4&NbIdent=1&Radius=2&Radius.unit=arcmin&submit=submit+id G055.7+3.4.], which is a supernova remnant. The data were taken on August 23, 2010 in the first D-configuration for which the new wideband capabilities of the WIDAR (Wideband Interferometric Digital ARchitecture) correlator were available. The 8-hour long session | We will be utilizing data taken, with the Karl G. Jansky Very Large Array, of [http://simbad.u-strasbg.fr/simbad/sim-id?Ident=SNR+G055.7%2B03.4&NbIdent=1&Radius=2&Radius.unit=arcmin&submit=submit+id G055.7+3.4.], which is a supernova remnant. The data were taken on August 23, 2010 in the first D-configuration for which the new wideband capabilities of the WIDAR (Wideband Interferometric Digital ARchitecture) correlator were available. The 8-hour long session included all available 1 GHz of bandwidth in L-band, from 1–2 GHz in frequency., although in this guide we will use dataset composed of only 4 spectral windows (128 MHz bandwidth each). | ||
The guide will often reference the CASA documentation archives which are available [https://casa.nrao.edu/casadocs/casa-5.4.0 online]. | The guide will often reference the CASA documentation archives which are available [https://casa.nrao.edu/casadocs/casa-5.4.0 online]. | ||
| Line 72: | Line 72: | ||
This guide has been written for CASA version 5.4.0. Please confirm your version before proceeding by checking the message in the command line interface window or the CASA logger after startup. | This guide has been written for CASA version 5.4.0. Please confirm your version before proceeding by checking the message in the command line interface window or the CASA logger after startup. | ||
For this tutorial, we will be running tasks using the | For this tutorial, we will be running tasks using the ''task (parameter = value)'' syntax. All parameters not explicitly called by this manner will use their default values. | ||
</div> | </div> | ||
| Line 212: | Line 212: | ||
It's important to be aware that the antennas have both a name and an ID associated with them. For example antenna ID 15 is named ea19 (the ''ea'' stemming from the Expanded VLA project). When specifying an antenna within a task parameter, we will primarily reference them by their ''ea'' name. | It's important to be aware that the antennas have both a name and an ID associated with them. For example antenna ID 15 is named ea19 (the ''ea'' stemming from the Expanded VLA project). When specifying an antenna within a task parameter, we will primarily reference them by their ''ea'' name. | ||
We can see the antenna configuration for this observing session by using [https://casa.nrao.edu/casadocs/casa-5. | We can see the antenna configuration for this observing session by using [https://casa.nrao.edu/casadocs/casa-5.4.0/data-examination-and-editing/plotting-antenna-positions plotants]. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 218: | Line 218: | ||
plotants(vis='SNR_G55_10s.ms', figfile='SNR_G55_10s.plotants.png') | plotants(vis='SNR_G55_10s.ms', figfile='SNR_G55_10s.plotants.png') | ||
</source> | </source> | ||
Figure 1 the antenna configuration for our observations; antennas ea01, ea03, and ea18 were on the extreme ends of the west, east, and north arms, respectively. | |||
<!-- This is really not for this guide, so commented out for the time being. ADK, Oct 2018--> | <!-- This is really not for this guide, so commented out for the time being. ADK, Oct 2018--> | ||
| Line 224: | Line 226: | ||
=== Identifying RFI with Plotms() === | === Identifying RFI with Plotms() === | ||
[[Image:CASA5.4.0-FLAG-Plotms-amp-time.png|300px|thumb|right|'''Figure 2''' <br />Plotms image of supernova remnant data, showing strong RFI through the observation (x axis) and affecting a number of spectral windows (data are colorised by spw).]] | |||
The data can be inspected with the [https://casa.nrao.edu/casadocs/casa-5. | The data can be inspected with the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms] task. | ||
It is worth first having a look at the data, separately per each field, in <b>amplitude vs. frequency</b> (to identify narrow-band RFI) and <b>amplitude vs. time</b> (intermittent RFI). | It is worth first having a look at the data, separately per each field, in <b>amplitude vs. frequency</b> (to identify narrow-band RFI) and <b>amplitude vs. time</b> (intermittent RFI). | ||
| Line 233: | Line 236: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='SNR_G55_10s.ms', field='1', xaxis='time', yaxis='amp', coloraxis=' | plotms(vis='SNR_G55_10s.ms', field='1', xaxis='time', yaxis='amp', coloraxis='spw') | ||
</source> | </source> | ||
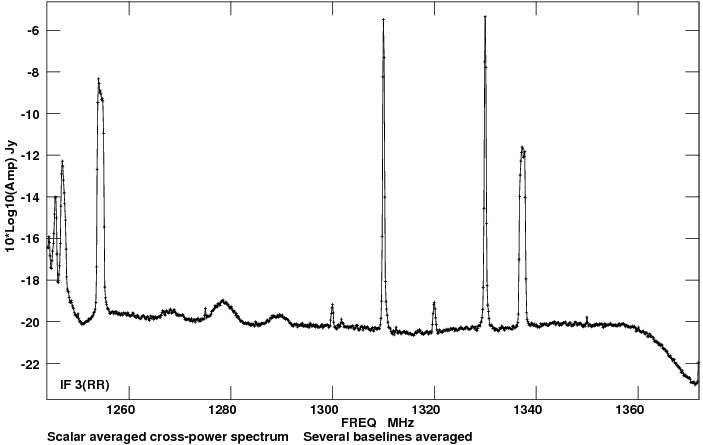
We can immediately see that there is plenty of RFI throughout the observations, affecting a number of spectral windows (data are colorised by spectral window; Figure 2). The RFI is discerned by the large spikes in amplitude. | |||
To narrow down where the RFI occurs let's now have a look at the data in amplitude vs frequency, plotted separately per each scan: | |||
<source lang="python"> | <source lang="python"> | ||
| Line 246: | Line 248: | ||
yaxis='amp', coloraxis='spw', iteraxis='scan') | yaxis='amp', coloraxis='spw', iteraxis='scan') | ||
</source> | </source> | ||
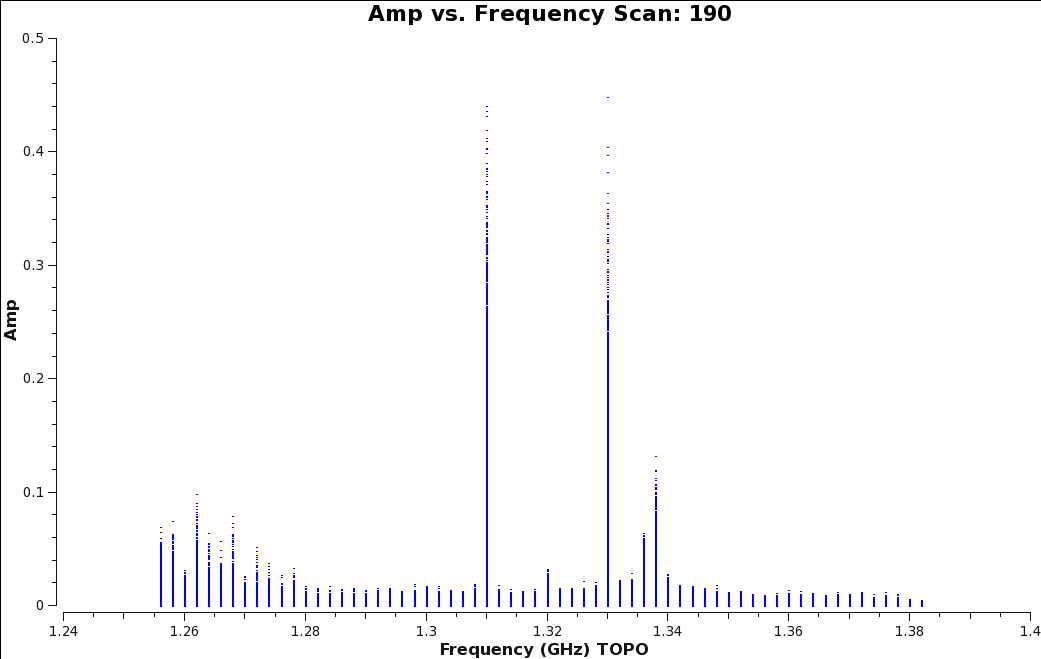
[[Image:amp_v_freq_rawdata.png|300px|thumb|right|'''Figure | [[Image:amp_v_freq_rawdata.png|300px|thumb|right|'''Figure 3''' <br />Plotms image of scan 190 showing strong RFI spikes in amplitude for several spectral windows.]] | ||
* coloraxis='spw': Parameter indicates that a different color will be assigned to each spectral window. | * coloraxis='spw': Parameter indicates that a different color will be assigned to each spectral window. | ||
* antenna='ea24' : Here we chose only information for antenna ea24. | * antenna='ea24' : Here we chose only information for antenna ea24. | ||
* iteraxis='scan': Parameter tells plotms to iterate over scans, that is to display a new plot for each scan. | * iteraxis='scan': Parameter tells plotms to iterate over scans, that is to display a new plot for each scan. | ||
The command above will set the plotms | The command above will set the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms] for iteration, but if you want to set this option from within [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms], go onto the Plot tab (horizontal), and then Page tab (vertical). You will now find Iteration options. Set Axis to the parameter you want to iterate through. Iterate by clicking on the green arrow buttons on the toolbar within the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms]. | ||
Progressing through to scan 190 , we can see there is significant time and frequency variable RFI present in the observing session | Progressing through to scan 190 , we can see there is significant time and frequency variable RFI present in the observing session. To determine which spectral windows are affected, click on the <b>Mark Regions</b> tool at the bottom of the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms] GUI (the open box with a green plus sign) and use the mouse to select a few of the highest-amplitude points in each of the spectral windows (Figure 3). Click on the <b>Locate</b> button (magnifying glass on white background). Information about the selected areas should now display in the logger window. For example: | ||
<br /> | <br /> | ||
</div> | </div> | ||
| Line 298: | Line 300: | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:RFI_1244-1372.png|380px|thumb|right|'''Figure | | [[Image:RFI_1244-1372.png|380px|thumb|right|'''Figure 4a''' <br />Amplitude vs. frequency plot of a portion of the L-band spectrum, showing RFI spikes from different sources.]] | ||
| [[Image:amp_v_freq.spw0_Scan190.png|330px|thumb|right|'''Figure | | [[Image:amp_v_freq.spw0_Scan190.png|330px|thumb|right|'''Figure 4b''' <br />Amplitude vs. frequency plot of L-Band, spectral window 0, for scan 190. Generated by running the command "plotms(vis='SNR_G55_10s.ms', spw='0', scan='190', xaxis='freq', yaxis='amp')" within CASA.]] | ||
|} | |} | ||
Reviewing the log output, we see that spw 0 and 2 are affected the most by RFI. We also get the corresponding frequency where the RFI is present, which appears to be 1.31 and 1.33 GHz for spectral window 0, and 1.686 GHz for spectral window 2. This information is important, especially when flagging data interactively as we will be doing towards the end of this guide. | Reviewing the log output, we see that spw 0 and 2 are affected the most by RFI. We also get the corresponding frequency where the RFI is present, which appears to be 1.31 and 1.33 GHz for spectral window 0, and 1.686 GHz for spectral window 2. This information is important, especially when flagging data interactively as we will be doing towards the end of this guide. | ||
The VLA has obtained RFI sweeps over its full frequency range. The results are available on the [https://science.nrao.edu/facilities/vla/docs/manuals/obsguide/modes/rfi Radio Frequency Interference website.] Most of the RFI features can be identified as radar, communications, satellites, airplanes, or birdies (i.e., RFI generated by the VLA electronics itself). For our observations, we inspect the L-band specific RFI plots [https://science.nrao.edu/facilities/vla/observing/RFI/L-Band website] and find that the 1310 and 1330 MHz features are due to FAA ASR radars, while the one at 1686 MHz is most likely due to a GOES weather satellite. We can compare the plots of the FAA ASR radar from the website (Figure | The VLA has obtained RFI sweeps over its full frequency range. The results are available on the [https://science.nrao.edu/facilities/vla/docs/manuals/obsguide/modes/rfi Radio Frequency Interference website.] Most of the RFI features can be identified as radar, communications, satellites, airplanes, or birdies (i.e., RFI generated by the VLA electronics itself). For our observations, we inspect the L-band specific RFI plots [https://science.nrao.edu/facilities/vla/observing/RFI/L-Band website] and find that the 1310 and 1330 MHz features are due to FAA ASR radars, while the one at 1686 MHz is most likely due to a GOES weather satellite. We can compare the plots of the FAA ASR radar from the website (Figure 4a), with that of spw 0 (Figure 4b), and see that the spikes are practically identical and fall within the same frequencies. | ||
=== Identifying RFI with MSview() === | === Identifying RFI with MSview() === | ||
| Line 311: | Line 312: | ||
Similar to using plotms() for the ms file editing, one can also use the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()] task. | Similar to using plotms() for the ms file editing, one can also use the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()] task. | ||
MSview()displays data in a 3D fashion, where the intensity of the plotted data points represents amplitude, phase, or their derivatives, while x and y axes of the display can be set to any of the following: time, baseline, channel, correlation or spectral window. For the introduction to the msview() tool see its dedicated [https://casaguides.nrao.edu/index.php/Data_flagging_with_viewer guide] (note that msview() can also be invoked as viewer(), as it is done in the linked guide). | MSview() displays data in a 3D fashion, where the intensity of the plotted data points represents amplitude, phase, or their derivatives, while x and y axes of the display can be set to any of the following: time, baseline, channel, correlation or spectral window. For the introduction to the msview() tool see its dedicated [https://casaguides.nrao.edu/index.php/Data_flagging_with_viewer guide] (note that msview() can also be invoked as viewer(), as it is done in the linked guide). | ||
[[Image:CASA5.4.0-flagging-msview-load-data.png|300px|thumb|right|'''Figure | [[Image:CASA5.4.0-flagging-msview-load-data.png|300px|thumb|right|'''Figure 5''' <br /> Loading data into msview(). The red box marks where data preselection can be made.]] | ||
<b>Note:</b> msview() will not handle large data volumes very well, and it may crash if your ms file is too large. If that happens try preselecting a fraction of your data for viewing. Also, sometimes restarting CASA may help. | <b>Note:</b> msview() will not handle large data volumes very well, and it may crash if your ms file is too large. If that happens try preselecting a fraction of your data for viewing. Also, sometimes restarting CASA may help. | ||
Since the task does not allow for much data selection in the command line, open msview() tool first, and load and select the data from its graphical interface: | Since the task does not allow for much data selection in the command line, open [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()] tool first, and load and select the data from its graphical interface: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
msview() | msview() | ||
</source> | </source> | ||
Once the tool opens, it will prompt you for data upload. At the same time as selecting the ms file to open, choose which field, spw etc you would like to load from that ms file as shown in Figure | Once the tool opens, it will prompt you for data upload. At the same time as selecting the ms file to open, choose which field, spw etc you would like to load from that ms file as shown in Figure 5, and hit Raster Image button. Only the preselected data will be available in the viewer, and further selection for viewing can be done via Data Display Options window (wrench button, within [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()]) under its ''display axes'' tab. Note that the ''ms and visibility selection'' tab will inform you about the overall shape of your data, even if you have not loaded the full ms file. | ||
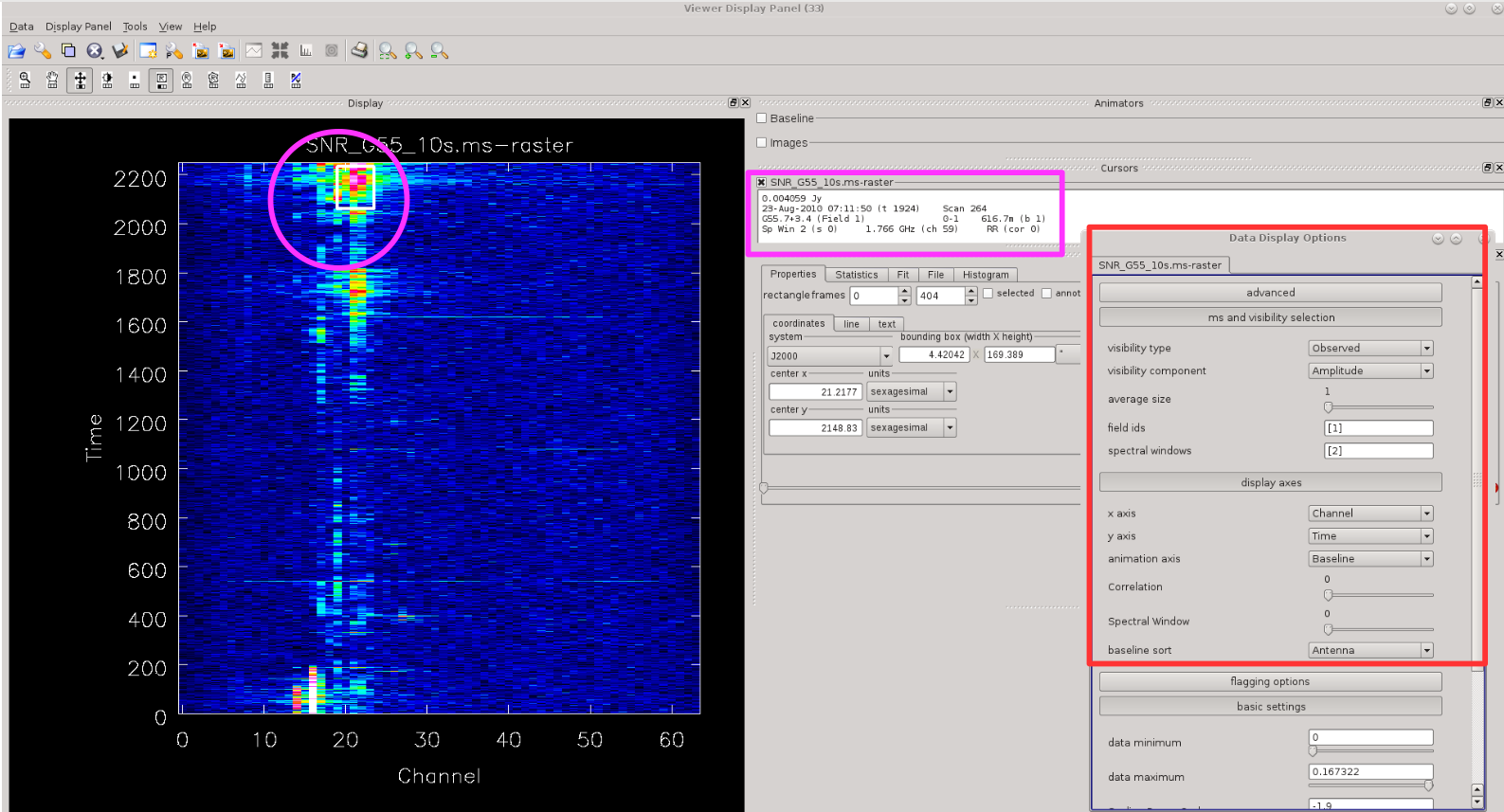
To identify RFI in msview(), one can look at the data in multiple ways. For example, let's plot <b>time vs. channel</b> for a single spectral window. We can set these selections in the Data Display Options (click on the Wrench button) as shown in Figure | To identify RFI in [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()], one can look at the data in multiple ways. For example, let's plot <b>time vs. channel</b> for a single spectral window. We can set these selections in the Data Display Options (click on the Wrench button) as shown in Figure 6. The selection window will appear (highlighted with red box in Figure 6). We plotted here spw=2, which corresponds to the orange-coloured spw from Figure 3. The blue coloured pixels are fairly well behaved data, while the green and red pixels are the data affected by the RFI. We can narrow down where that RFI occurs by hovering cursor over the RFI areas (white box/magenta circle in Figure 6) and details on affected baselines, scans, etc will be listed in the Cursors window (highlighted with magenta box in Figure 6). Note that [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview] refers to the antennas via their ID numbers and not the ''ea'' names (compare with the listobs output and commentary in section [[#Preliminary_Data_Evaluation | Preliminary Data Evaluation]]). Also, note that here only RR polarisation is plotted, and you will need to check also LL (as well as RL and LR if you have and use them in further data processing). | ||
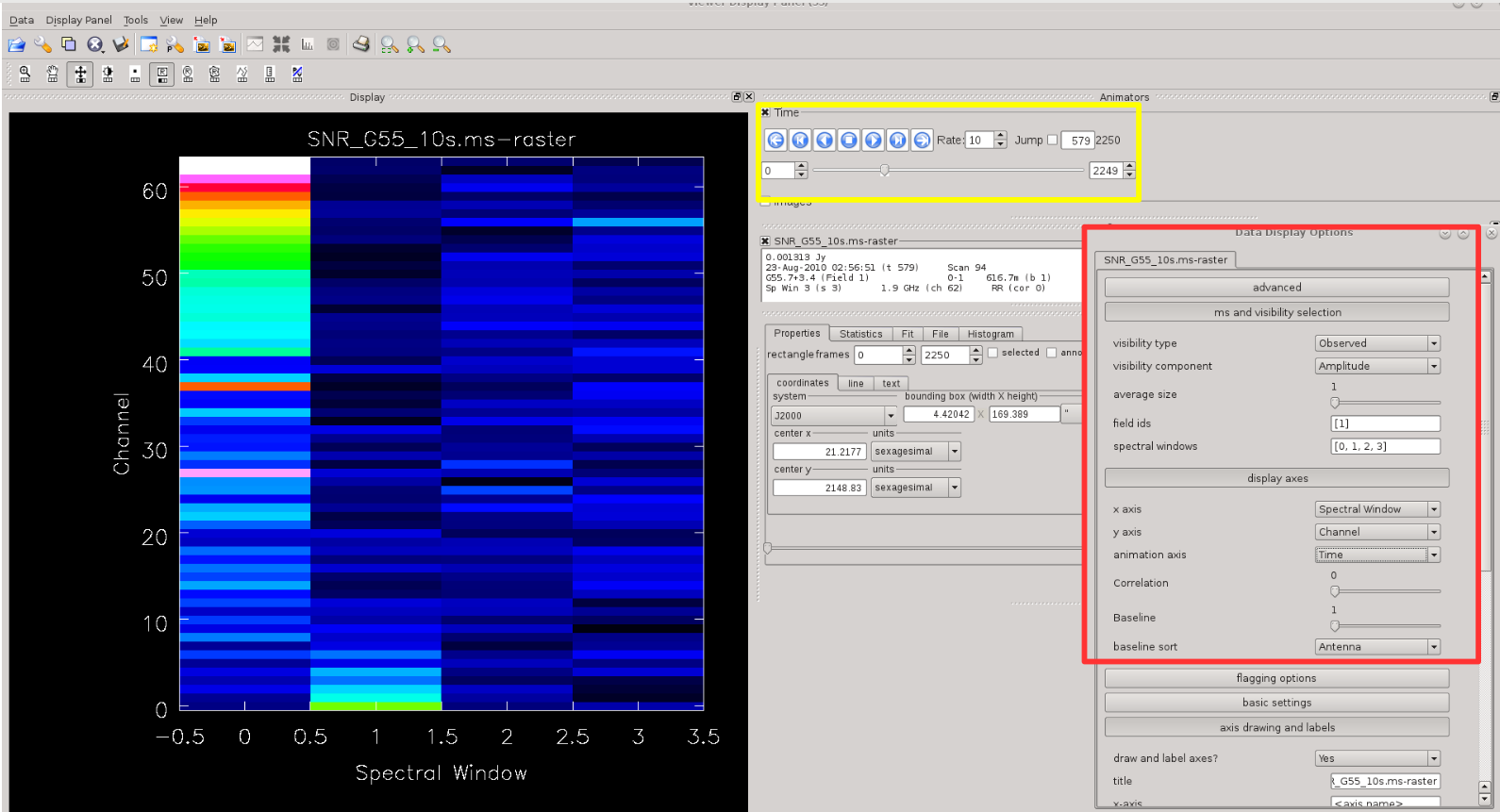
One can also have a quick look at which spectral windows most RFI occurs, and when. For example, let's plot <b>channel vs. spectral window</b> and iterate over time Figure | One can also have a quick look at which spectral windows most RFI occurs, and when. For example, let's plot <b>channel vs. spectral window</b> and iterate over time Figure 7). You can choose iteration by selecting animation axis under the ''display axes'' tab within the Data Display Options (marked with red box in Figure 7). To inspect how the RFI changes with time use the iteration buttons within the Animators panel (marked with yellow box in Figure 7). Again, the blue colour marks the well behaved data here; you can change the colour scheme in the Data Display Option window. | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:CASA5.4.0-flagging-Msview-time-chan-spw2.png| | | [[Image:CASA5.4.0-flagging-Msview-time-chan-spw2.png|400px|thumb|right|'''Figure 6''' <br /> Time vs channel plot visualised with msview(). The data selection window for viewing is highlighted with the red box. The magenta box gives information on pixels which we hovered over with a cursor (white box/magenta circle) and which are affected by RFI.]] | ||
| [[Image:CASA5.4.0-flagging-Msview-chan-spw-iterate-time.png| | | [[Image:CASA5.4.0-flagging-Msview-chan-spw-iterate-time.png|400px|thumb|right|'''Figure 7''' <br /> Channel vs spectral window plotted with msview(). The data selection window for viewing is highlighted with the red box. The yellow box indicates buttons to play a movie over the iteration axis, which here is set to time.]] | ||
|} | |} | ||
| Line 339: | Line 339: | ||
We first check the operator log for the observing session to see if there were any issues noted during the run that need to be addressed. The logs are available from the [http://www.vla.nrao.edu/cgi-bin/oplogs.cgi VLA operator log website]. Here is the link for the [http://www.vla.nrao.edu/operators/logs/2010/8/2010-08-23_0005_AB1345.pdf observing log] file for our observing session. | We first check the operator log for the observing session to see if there were any issues noted during the run that need to be addressed. The logs are available from the [http://www.vla.nrao.edu/cgi-bin/oplogs.cgi VLA operator log website]. Here is the link for the [http://www.vla.nrao.edu/operators/logs/2010/8/2010-08-23_0005_AB1345.pdf observing log] file for our observing session. | ||
The log has various pieces of information including the start/end times for the observing session, frequency bands used, weather, baseline information for recently moved antennas, and any outages or issues that may have been encountered. We see that antenna ea07 may need position corrections (see one of our calibration tutorials, [https://casaguides.nrao.edu/index.php?title=VLA_high_frequency_Spectral_Line_tutorial_-_IRC%2B10216-CASA5. | The log has various pieces of information including the start/end times for the observing session, frequency bands used, weather, baseline information for recently moved antennas, and any outages or issues that may have been encountered. We see that antenna ea07 may need position corrections (see one of our calibration tutorials, [https://casaguides.nrao.edu/index.php?title=VLA_high_frequency_Spectral_Line_tutorial_-_IRC%2B10216-CASA5.4.0 IRC+10216], on how to fix this), and several antennas (ea06, ea17, ea20, and ea26) are missing an L-Band receiver. Antenna 07 position corrections will need to be applied during data calibration and will not be covered in this tutorial. Additionally, the antennas with missing receivers were already removed from the dataset used here (as explained in the above section Obtaining the Data), so we can continue with online flagging. | ||
<br /> | <br /> | ||
<br /> | <br /> | ||
| Line 350: | Line 350: | ||
We will now apply these online flags to the data by employing [https://casa.nrao.edu/casadocs/casa-5.4.0/data-examination-and-editing/flagging-based-on-a-list-of-commands-flagcmd flagcmd], but first, let's create a plot of the flags to get an idea of what will be flagged. | We will now apply these online flags to the data by employing [https://casa.nrao.edu/casadocs/casa-5.4.0/data-examination-and-editing/flagging-based-on-a-list-of-commands-flagcmd flagcmd], but first, let's create a plot of the flags to get an idea of what will be flagged. | ||
[[Image:flaggingreason_vs_time.png|300px|thumb|right|'''Figure | [[Image:flaggingreason_vs_time.png|300px|thumb|right|'''Figure 8''' Online Flags]] | ||
<source lang="python"> | <source lang="python"> | ||
| Line 357: | Line 357: | ||
</source> | </source> | ||
Open the ''flaggingreason_vs_time.png'' with your favorite image viewing program. Figure | Open the ''flaggingreason_vs_time.png'' with your favorite image viewing program. Figure 8 shows several instances of online flagging. Most notably, ea28 and ea08 had some subreflector issues throughout the observing session. Online flags are instances of possible missing data, including: | ||
* ANTENNA_NOT_ON_SOURCE | * ANTENNA_NOT_ON_SOURCE | ||
| Line 392: | Line 392: | ||
</source> | </source> | ||
[[Image:Elevation_vs_Time.png|300px|thumb|right|'''Figure | [[Image:Elevation_vs_Time.png|300px|thumb|right|'''Figure 9''' Plot of elevation vs. time.]] | ||
Figure | Figure 9 shows that most of the sources were at or above 40 degrees in elevation; accordingly, we expect a minimal amount of shadowing. The flux density / bandpass calibrator 3C147 was observed towards the end of the session, which could result in some shadowing of antennas due to its low elevation of about 20–22 degrees. | ||
Task [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] can determine and flag shadowed antennas by their location and observing direction: | Task [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] can determine and flag shadowed antennas by their location and observing direction: | ||
| Line 466: | Line 466: | ||
<b>Note: As Hanning-smoothing will remove amplitude spikes, it is not recommended for spectral analysis related science (e.g. strong narrow maser lines).</b> | <b>Note: As Hanning-smoothing will remove amplitude spikes, it is not recommended for spectral analysis related science (e.g. strong narrow maser lines).</b> | ||
Let's create before (Figure | Let's create before (Figure 10a) and after images (Figure 10b) with the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms] task to see the effect. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 490: | Line 490: | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:amp_v_freq.beforeHanning.png|350px|thumb|right|'''Figure | | [[Image:amp_v_freq.beforeHanning.png|350px|thumb|right|'''Figure 10a''' <br />Plot of amplitude vs. frequency before Hanning smoothing data.]] | ||
| [[Image:amp_v_freq.afterHanning.png|350px|thumb|right|'''Figure | | [[Image:amp_v_freq.afterHanning.png|350px|thumb|right|'''Figure 10b''' <br />Plot of amplitude vs. frequency after Hanning smoothing data.]] | ||
|} | |} | ||
Figure | Figure 10b shows the effect of applying Hanning-smoothing. Notice that single channel RFI has spread into three channels. This spreading of RFI to other channels will ultimately result in a little more data being flagged. | ||
== Automatic RFI Excision == | == Automatic RFI Excision == | ||
| Line 509: | Line 509: | ||
Step 4: Search for low-level wings of very strong RFI. <br /> | Step 4: Search for low-level wings of very strong RFI. <br /> | ||
We will apply the auto-flagging TFCrop algorithm for each spectral window. With TFCrop, it's a good idea to first inspect a small portion of data and review what and how much will be flagged. Once you are comfortable with the results, you can apply the algorithm to the remaining data. We will walk through the first spw and then include the remaining spws with the same command. First plot the corrected data for scan 190 (Figure | [[Image:amp_v_freq_before_tfcrop.png|350px|thumb|right|'''Figure 7''' <br />Amplitude vs. Frequency plot of Scan 190, before the TFCrop algorithm is applied.]] | ||
We will apply the auto-flagging TFCrop algorithm for each spectral window. With TFCrop, it's a good idea to first inspect a small portion of data and review what and how much will be flagged. Once you are comfortable with the results, you can apply the algorithm to the remaining data. We will walk through the first spw and then include the remaining spws with the same command. First plot the corrected data for scan 190 (Figure 11) with all spectral windows so we can compare the data before and after TFCrop. | |||
<source lang="python"> | <source lang="python"> | ||
| Line 519: | Line 520: | ||
The following are a set of [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] commands which have been found to work reasonably well with these data. Take some time to play with the parameters, and the plotting capabilities, as this will be important when you are working with your own data later on. Because we set parameters ''display='both' ''and ''action='calculate', '' the flags are displayed but not actually written to the MS. This allows one to try different sets of parameters before actually applying the flags to the data. | The following are a set of [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] commands which have been found to work reasonably well with these data. Take some time to play with the parameters, and the plotting capabilities, as this will be important when you are working with your own data later on. Because we set parameters ''display='both' ''and ''action='calculate', '' the flags are displayed but not actually written to the MS. This allows one to try different sets of parameters before actually applying the flags to the data. | ||
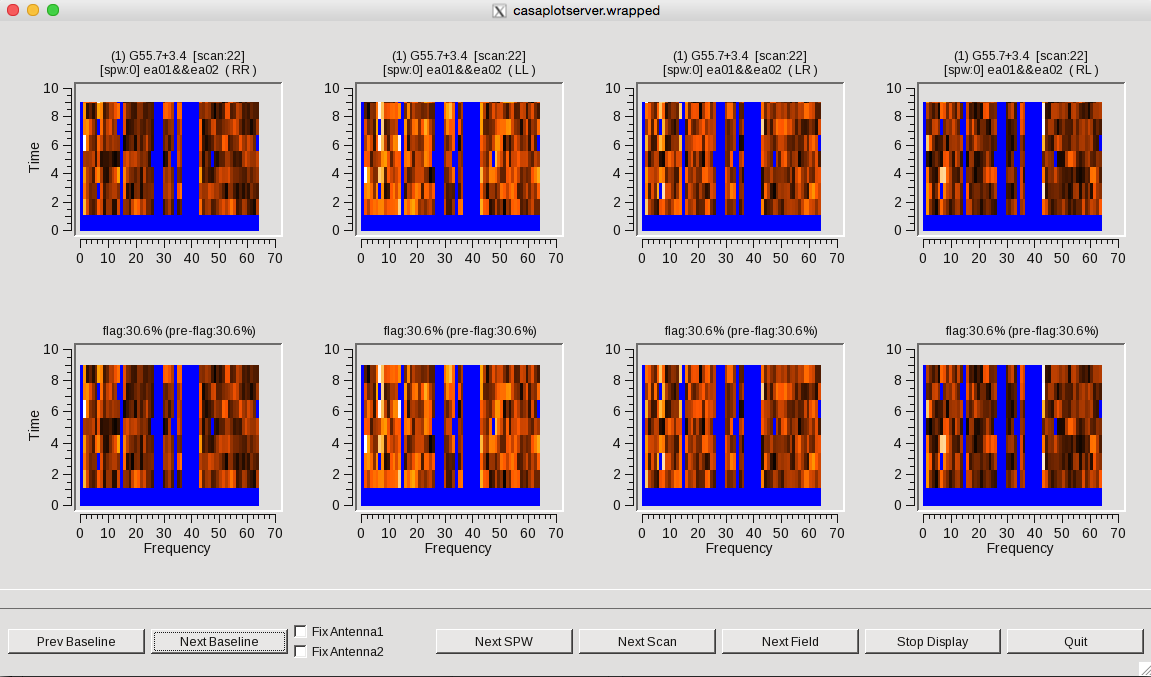
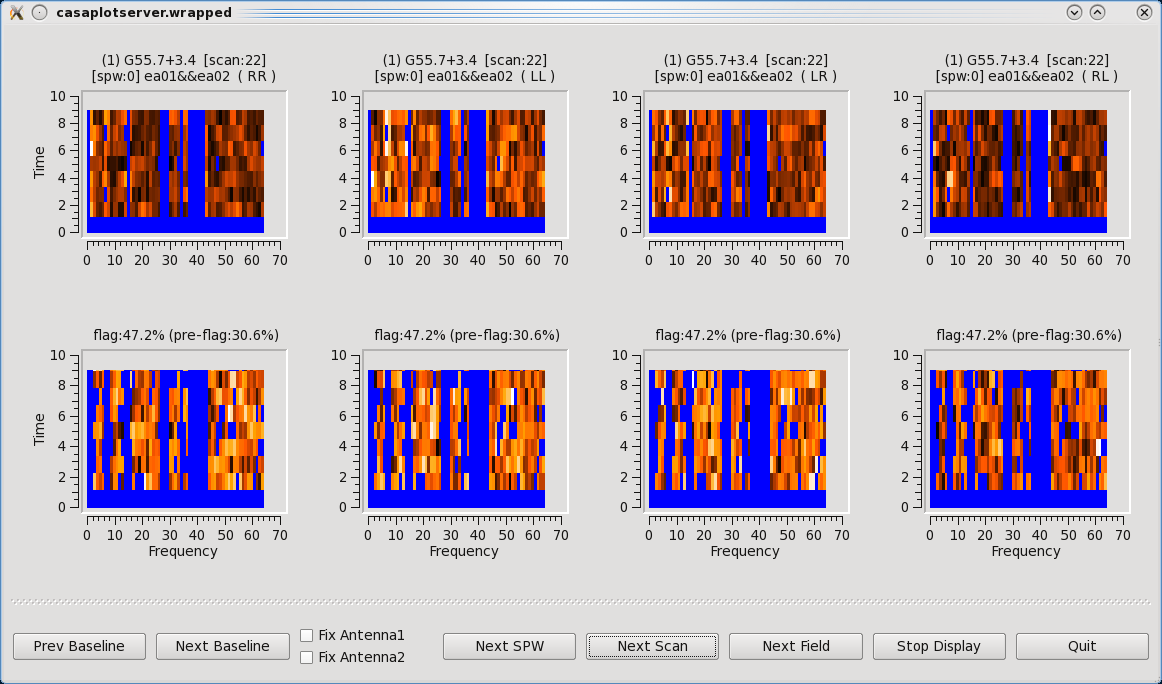
[[Image:Tfcrop.png|350px|thumb|right|'''Figure 12''' <br />TFCrop flagging results. Top row is before, and bottom row is after TFCrop has applied flagging.]] | |||
Some representative TFCrop plots are displayed (Figure | Some representative TFCrop plots are displayed (Figure 12). Each column displays an individual polarization product; all four polarizations are, from left to right, RR, RL, LR, and LL. The first row shows the data with current flags applied and the second includes the flags generated by flagdata. The x-axis is channel number (the spectral window ID is displayed in the top title) and the y-axis of the first two rows is all integrations included in a time segment, set by the parameter ''ntime''. These are the data considered by the TFCrop algorithm during its flagging process, and changes in ''ntime'' will have some (relatively small) effect on what data are flagged. | ||
Each plot page displays data for a single baseline and time segment, with colours representing the intensity of the visibilities (amplitude) in the heat colour scheme ranging from black for lowest amplitude points to white for highest amplitude data points. The flagged, or to be flagged, visibilities are masked in blue. The buttons at the bottom allow one to step through baseline (backward and forward), spw, scan, and field while Stop Display will continue the flagging operation without the GUI, and Quit aborts the run. | Each plot page displays data for a single baseline and time segment, with colours representing the intensity of the visibilities (amplitude) in the heat colour scheme ranging from black for lowest amplitude points to white for highest amplitude data points. The flagged, or to be flagged, visibilities are masked in blue. The buttons at the bottom allow one to step through baseline (backward and forward), spw, scan, and field while Stop Display will continue the flagging operation without the GUI, and Quit aborts the run. | ||
| Line 532: | Line 534: | ||
display='both', flagbackup=False) | display='both', flagbackup=False) | ||
</source> | </source> | ||
The first two scans (14,15) are fully flagged (hence displayed as completely blue). Click on Next Scan until you reach scan 17 where it will be easier to see the flagging the algorithm performs. The top row shows current flags already applied to the data. The bottom row illustrates the flagging that can be applied with TFCrop, represented by the additional blue areas. Iterate through several scans and baselines. Once you're done, click on the Quit button on the bottom right corner. Let's now apply these flags by changing the action parameter. | The first two scans (14,15) are fully flagged (hence displayed as completely blue). Click on Next Scan until you reach scan 17 where it will be easier to see the flagging the algorithm performs. The top row shows current flags already applied to the data. The bottom row illustrates the flagging that can be applied with TFCrop, represented by the additional blue areas. Iterate through several scans and baselines. Once you're done, click on the Quit button on the bottom right corner. Let's now apply these flags by changing the action parameter. | ||
| Line 547: | Line 547: | ||
''' spw 1, 2, 3 ''' | ''' spw 1, 2, 3 ''' | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
| Line 555: | Line 556: | ||
Iterate through several scans and baselines and, once you're ready to apply the flags, click on Stop Display. After TFCrop has gone through and flagged some of the worst RFI, we can inspect the log report and take note of how much has been flagged. | Iterate through several scans and baselines and, once you're ready to apply the flags, click on Stop Display. After TFCrop has gone through and flagged some of the worst RFI, we can inspect the log report and take note of how much has been flagged. | ||
We can also use [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms] to review the effects of using TFCrop | We can also use [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms] to review the effects of using TFCrop. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 563: | Line 564: | ||
correlation='RR,LL', coloraxis='spw', plotrange=[1.2,2,-0.01,0.25]) | correlation='RR,LL', coloraxis='spw', plotrange=[1.2,2,-0.01,0.25]) | ||
</source> | </source> | ||
Compare the plots before applying TFCrop (Figure 11) to the plot after applying TFCrop (Figure 13). Figure 13 shows great improvements, especially for spectral window 0 and 2, which had some of the worst RFI. | |||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:amp_v_freq_before_tfcrop.png|350px|thumb|right|'''Figure | | [[Image:amp_v_freq_before_tfcrop.png|350px|thumb|right|'''Figure 11''' <br />Amplitude vs. Frequency plot of Scan 190, before the TFCrop algorithm is applied.]] | ||
| [[Image:amp_v_freq_after_tfcrop_Scan190.png|350px|thumb|right|'''Figure | | [[Image:amp_v_freq_after_tfcrop_Scan190.png|350px|thumb|right|'''Figure 13''' <br />Plot of amplitude vs. frequency showing the effects of applying TFCrop to our data, for scan 190.]] | ||
|} | |} | ||
''' Flagging Summary ''' | ''' Flagging Summary ''' | ||
We now calculate the amount of flagged | We now calculate the amount of data flagged so far in our Measurement Set by using the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] task with parameter ''mode='summary' ''and assign the returned Python dictionary to the variable flagInfo. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 617: | Line 620: | ||
==== Bandpass calibration (to demonstrate RFlag) ==== | ==== Bandpass calibration (to demonstrate RFlag) ==== | ||
For the purpose of demonstrating the functionality of RFlag on our data, we need to perform bandpass calibration first. Here, we just briefly outline the bandpass calibration process. For detailed calibration tutorial see [https://casaguides.nrao.edu/index.php/Karl_G._Jansky_VLA_Tutorials#VLA_Data_Reduction_Tutorials | For the purpose of demonstrating the functionality of RFlag on our data, we need to perform bandpass calibration first. Here, we just briefly outline the bandpass calibration process. For detailed calibration tutorial see [https://casaguides.nrao.edu/index.php/Karl_G._Jansky_VLA_Tutorials#VLA_Data_Reduction_Tutorials VLA Data Reduction Tutorials]. | ||
First, | First, run [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_gaincal/about gaincal] to solve for an initial set of antenna-based phases over a narrow range of channels. To find channels for each spectral window that are relatively RFI-free over the course of the observing session, look at the data with [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms]. Use only a narrow channel range. | ||
<!-- | <!-- | ||
THIS IS NOT NEEDED FOR THIS FLAGGING TUTORIAL, REF TO CALIBRATION TUTORIALS GIVEN | THIS IS NOT NEEDED FOR THIS FLAGGING TUTORIAL, REF TO CALIBRATION TUTORIALS GIVEN | ||
| Line 632: | Line 635: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='SNR_G55_10s-hanning.ms', scan='42,65,88,11,134,157', antenna='ea24', xaxis='channel', yaxis='amp', iteraxis='spw') | plotms(vis='SNR_G55_10s-hanning.ms', scan='42,65,88,11,134,157', antenna='ea24', xaxis='channel', | ||
yaxis='amp', iteraxis='spw') | |||
</source> | </source> | ||
Iterating over each spectral window plot, search for channel ranges with stable amplitudes. An example of a few appropriate channel ranges include: SPW 0:20~24, SPW 1:49~52, SPW 2:38~41, SPW 3:41~44. | Iterating over each spectral window plot, search for channel ranges with stable amplitudes. An example of a few appropriate channel ranges include: SPW 0:20~24, SPW 1:49~52, SPW 2:38~41, SPW 3:41~44. | ||
Using these ranges, | Using these ranges, run [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_gaincal/about gaincal] to calculate phase-only solutions that will be used as input during the initial bandpass calibration. Remember: the calibration tables we are creating now are so that we can use an automatic RFI flagging algorithm. The final calibration tables will be generated later, after automated flagging. Here are the inputs for the initial pre-bandpass phase calibration: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
gaincal(vis='SNR_G55_10s-hanning.ms', caltable='SNR_G55_10s-hanning.initPh', field='J1925+2106', solint=' int ', spw='0:20~24,1:49~52,2:38~41,3:41~44', refant='ea24', minblperant=3, minsnr=3.0, calmode='p') | gaincal(vis='SNR_G55_10s-hanning.ms', caltable='SNR_G55_10s-hanning.initPh', field='J1925+2106', | ||
solint=' int ', spw='0:20~24,1:49~52,2:38~41,3:41~44', refant='ea24', minblperant=3, | |||
minsnr=3.0, calmode='p') | |||
</source> | </source> | ||
<!-- | <!-- | ||
| Line 657: | Line 663: | ||
Note that a number of solutions do not pass the requirements of the minimum 3 baselines (generating the terminal message''' "Insufficient unflagged antennas to proceed with this solve."''') or minimum signal-to-noise ratio (outputting''' "n of x solutions rejected due to SNR being less than 3 ..."'''). The logger output indicates 320-324 solutions succeeded out of 387 attempted. | Note that a number of solutions do not pass the requirements of the minimum 3 baselines (generating the terminal message''' "Insufficient unflagged antennas to proceed with this solve."''') or minimum signal-to-noise ratio (outputting''' "n of x solutions rejected due to SNR being less than 3 ..."'''). The logger output indicates 320-324 solutions succeeded out of 387 attempted. | ||
[[Image:gain.phase_v_time.plotcal.png|300px|thumb|right|'''Figure 10a''' <br /> Gain Phase vs. Time for spw0, for four antennas.]] | |||
--> | --> | ||
[[Image: | [[Image:CASA5.4.0-Plotms-pahse-time.png|300px|thumb|right|'''Figure 14''' <br /> Gain Phase vs. Time for spw0, for four antennas.]] | ||
It | It is always a good idea to inspect the resulting calibration table. We can do this with [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms]. | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='SNR_G55_10s-hanning.initPh', xaxis='time', yaxis='phase', | |||
iteraxis='spw', antenna='ea01,ea05,ea10,ea24', coloraxis='Antenna1') | |||
</source> | </source> | ||
Figure | |||
Figure 14 shows the phases do not change much over the course of the observing session. | |||
<!-- | <!-- | ||
THIS IS NOT NEEDED FOR THIS FLAGGING TUTORIAL, REF TO CALIBRATION TUTORIALS GIVEN | THIS IS NOT NEEDED FOR THIS FLAGGING TUTORIAL, REF TO CALIBRATION TUTORIALS GIVEN | ||
| Line 674: | Line 683: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotcal(caltable='SNR_G55_10s-hanning.initPh', xaxis='time', yaxis='phase', iteration='spw', antenna='', plotsymbol = '-', plotrange=[-1,-1,-180,180]) | plotcal(caltable='SNR_G55_10s-hanning.initPh', xaxis='time', yaxis='phase', | ||
iteration='spw', antenna='', plotsymbol = '-', plotrange=[-1,-1,-180,180]) | |||
</source> | </source> | ||
| Line 684: | Line 694: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
bandpass(vis='SNR_G55_10s-hanning.ms', caltable='SNR_G55_10s-hanning.initBP', field='J1925+2106', solint=' inf ', combine='scan', refant='ea24', minblperant=3, minsnr=10.0, gaintable='SNR_G55_10s-hanning.initPh', interp='nearest', solnorm=False) | bandpass(vis='SNR_G55_10s-hanning.ms', caltable='SNR_G55_10s-hanning.initBP', | ||
field='J1925+2106', solint=' inf ', combine='scan', refant='ea24', | |||
minblperant=3, minsnr=10.0, gaintable='SNR_G55_10s-hanning.initPh', | |||
interp='nearest', solnorm=False) | |||
</source> | </source> | ||
<!-- | <!-- | ||
| Line 696: | Line 709: | ||
--> | --> | ||
Now, inspect the resulting bandpass plots with | Now, inspect the resulting bandpass plots with plotms (Figures 15a,b,c). | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='SNR_G55_10s-hanning.initBP',xaxis='freq', yaxis='amp', iteraxis='antenna', | |||
coloraxis='corr',gridrows=2) | |||
</source> | </source> | ||
{|style="margin: 0 auto;" | |||
--> | | [[Image:CASA5.4.0-Plotms-amp-freq-ea01-02.png|300px|thumb|right|'''Figure 15a''' <br /> Bandpass for antennas ea01 and ea02.]] | ||
| [[Image:CASA5.4.0-Plotms-amp-freq-ea03-04.png|300px|thumb|right|'''Figure 15b''' <br /> Bandpass for antennas ea03 and ea04.]] | |||
| [[Image:CASA5.4.0-Plotms-amp-freq-ea05-07.png|300px|thumb|right|'''Figure 15c''' <br /> Bandpass for antennas ea05 - ea07 (note ea06 is flagged).]] | |||
|} | |||
<br> | <br> | ||
</div> | </div> | ||
| Line 713: | Line 729: | ||
==== Interactive flagging of bad solutions (calibration tables) ==== | ==== Interactive flagging of bad solutions (calibration tables) ==== | ||
We notice that antennas ea01 and ea05 have a point that is offset from the rest ( | [[Image:CASA5.4.0-Plotms-amp-freq-ea01-05-flagging.png|300px|thumb|right|'''Figure 16''' <br /> Interactive plotms flagging of the offset points for ea01 and ea05 in the calibration table.]] | ||
We notice that antennas ea01 and ea05 have a point that is offset from the rest (Figures 15a,c). Let's plot just these two antennas, locate the point on the plot, and flag it interactively through [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plot]. The interactive flagging within plotms will not create a backup, therefore it may be wise to use [https://casa.nrao.edu/casadocs/casa-5.4.0/data-examination-and-editing/managing-flag-versions-flagmanager flagmanager] before doing so (see Section [[#Backup_Data_-_Flagmanager |Flagmanager]] earlier in this tutorial). | |||
<source lang="python"> | <source lang="python"> | ||
| Line 722: | Line 740: | ||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
plotms(vis='SNR_G55_10s-hanning.initBP',xaxis='freq', yaxis='amp', iteraxis='antenna', | |||
antenna='ea01,ea05', coloraxis='corr', gridrows=2) | |||
</source> | </source> | ||
We will highlight the points by clicking on the Mark Region button, drawing boxes over the points, and clicking on the Flag button. Before doing this, one could also get information on the points by clicking on the Locate button, which will display details about the highlighted regions (Figure | We will highlight the points by clicking on the <b>Mark Region</b> button, drawing boxes over the points, and clicking on the <b>Flag</b> button. Before doing this, one could also get information on the points by clicking on the Locate button, which will display details about the highlighted regions (Figure 16). After having flagged the offset points, your plots should update. | ||
<div style="background-color: #eeeeee; margin: 40px"> | <div style="background-color: #eeeeee; margin: 40px"> | ||
| Line 745: | Line 764: | ||
</div> | </div> | ||
</div> | </div> | ||
==== Running RFlag on bandpass calibrated data ==== | ==== Running RFlag on bandpass calibrated data ==== | ||
[[Image:amp_v_freq.beforeRFlag.png|320px|thumb|right|'''Figure 17''' <br />Amplitude vs. Frequency plot of scan 190, before RFlag is applied.]] | |||
Now that we have bandpass-corrected data, we will run [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata]] in rflag mode. | Now that we have bandpass-corrected data, we will run [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata]] in rflag mode. | ||
| Line 760: | Line 774: | ||
''' spw 0 ''' | ''' spw 0 ''' | ||
First, let's create a plot of our corrected data before we apply RFlag (Figure | First, let's create a plot of our corrected data before we apply RFlag (Figure 17). | ||
<source lang="python"> | <source lang="python"> | ||
| Line 776: | Line 790: | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:rflag_default_values.png| | | [[Image:rflag_default_values.png|400px|thumb|right|'''Figure 18''' <br />RFlag with Default Parameters for scan 22 (use the next scan, next baseline, and next field buttons to see this specific screen).]] | ||
| [[Image:rflag_freq2.5_time3.5.png| | | [[Image:rflag_freq2.5_time3.5.png|400px|thumb|right|'''Figure 19''' <br />RFlag with freqdevscale=2.5, and timedevscale=3.5 for scan 22. We can see more data are being flagged with these more stringent values.]] | ||
|} | |} | ||
After reviewing the calculated flags, we can see that a lot of RFI is being missed (Figure | After reviewing the calculated flags, we can see that a lot of RFI is being missed (Figure 18). We will need to modify the default parameters for this spectral window. Click on Quit on the lower right hand side of the window. Let's review the parameters for [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] and modify them: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 792: | Line 806: | ||
</source> | </source> | ||
The default timedevscale and freqdevscale | The default ''timedevscale=5.0'' and ''freqdevscale=5.0'' parameters are not flagging enough bad data. We can provide more stringent values to the time/freq deviation scales parameters. A value smaller than the default (5.0) will flag more data in either the time or frequency axes, as can be seen on the plots in the displayed window. A value larger than the default will result in less flagged data. | ||
Since we know spw 0 has lots of RFI, we will change parameters ''freqdevscale=2.5'' and ''timedevscale=3.5''. We will also change parameter ''action='apply'.'' Note that we can always decide that this still isn't good enough and click on <b>Quit</b> to change our parameter values (will not perform flagging). Once the window opens, | Since we know spw 0 has lots of RFI, we will change parameters ''freqdevscale=2.5'' and ''timedevscale=3.5''. We will also change parameter ''action='apply'.'' Note that we can always decide that this still isn't good enough and click on <b>Quit</b> to change our parameter values (will not perform flagging). Once the window opens, proceed to scan 22 (Figure 19), review what will be flagged, and click on <b>Stop Display</b> (as we did during TFCrop) to allow the task to continue with the flagging. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 858: | Line 872: | ||
</source> | </source> | ||
Let's create an after RFlag plot to see the improvements (Figure | Let's create an after RFlag plot to see the improvements (Figure 20). Compare the resulting plot to the before TFCrop plot (Figure 17). | ||
<source lang="python"> | <source lang="python"> | ||
| Line 868: | Line 882: | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:amp_v_freq.beforeRFlag.png|350px|thumb|right|'''Figure | | [[Image:amp_v_freq.beforeRFlag.png|350px|thumb|right|'''Figure 17''' <br />Amplitude vs. Frequency plot of scan 190, before RFlag is applied.]] | ||
| [[Image:amp_v_freq.before_after_RFlag.png|350px|thumb|right|'''Figure | | [[Image:amp_v_freq.before_after_RFlag.png|350px|thumb|right|'''Figure 20''' <br /> Plot of amplitude vs. frequency of scan 190, after having run RFlag.]] | ||
|} | |} | ||
== Interactive Flagging == | == Interactive Flagging == | ||
| Line 889: | Line 902: | ||
</source> | </source> | ||
Figure | Figure 21 (next section) shows some points in green that have higher amplitudes than the rest. Use the <b>Mark Regions</b> button (square with green plus sign) to highlight the region, and the <b>Locate</b> button (magnifying glass with white page) to obtain information on the data points. We can now decide to flag all of the highlighted area by clicking on the <b>Flag</b> button (red flag). The region is now flagged. | ||
Open | Similarly, one can also perform interactive flagging with [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview]. Again, the flagging is performed directly on the data, and so it is beneficial to execute flagmanager as above before embarking on interactive flagging with msview(). | ||
Open msview() and load the data as described in section [[#Identifying_RFI_with_MSview | Identifying RFI with MSview]]: | |||
<source lang="python"> | <source lang="python"> | ||
# In CASA | # In CASA | ||
msview() | msview() | ||
</source> | </source> | ||
Select what you would like the axes to display (by default msview will plot time vs baseline), and using zoom tool (magnifying glass button with plus sign) zoom in on the data chunks you would like to flag. Now, draw a region around the bad data using the <b>drawing selectors</b> (rectangle, circle, polygon) and <b>right-double click</b>. | Select what you would like the axes to display (by default msview will plot time vs baseline), and using zoom tool (magnifying glass button with plus sign) zoom in on the data chunks you would like to inspect and potentially flag. Now, draw a region around the bad data using the <b>drawing selectors</b> (rectangle, circle, polygon) and <b>right-double click</b> to flag. To save the flagging, click <b>Save Edits</b> button in the flagging tab of Data Display Option window. | ||
<pre> | <pre> | ||
| Line 907: | Line 920: | ||
</pre> | </pre> | ||
A more proper form of flagging would be to narrow your search on bad data points and use a flagging task to remove them as showed in the next section below. | A more proper form of flagging would be to narrow your search on bad data points (Locate data as described above) and use a flagging task to remove them as showed in the next section below. | ||
== Identifying remaining bad data with plotms()/msview() == | == Identifying remaining bad data with plotms()/msview() == | ||
Once the bulk of the RFI and corrupted data has been removed with the methods described earlier in this guide, it is worth having another look at the data with the visualisation tools, either plotms() or msview(). This step may be valuable to perform once your data are calibrated and ready for imaging, to double check all the bad data has been indeed removed. | Once the bulk of the RFI and corrupted data has been removed with the methods described earlier in this guide, it is worth having another look at the data with the visualisation tools, either [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_plotms/about plotms()] or [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()]. This step may be valuable to perform once your data are calibrated and ready for imaging, to double check all the bad data has been indeed removed. | ||
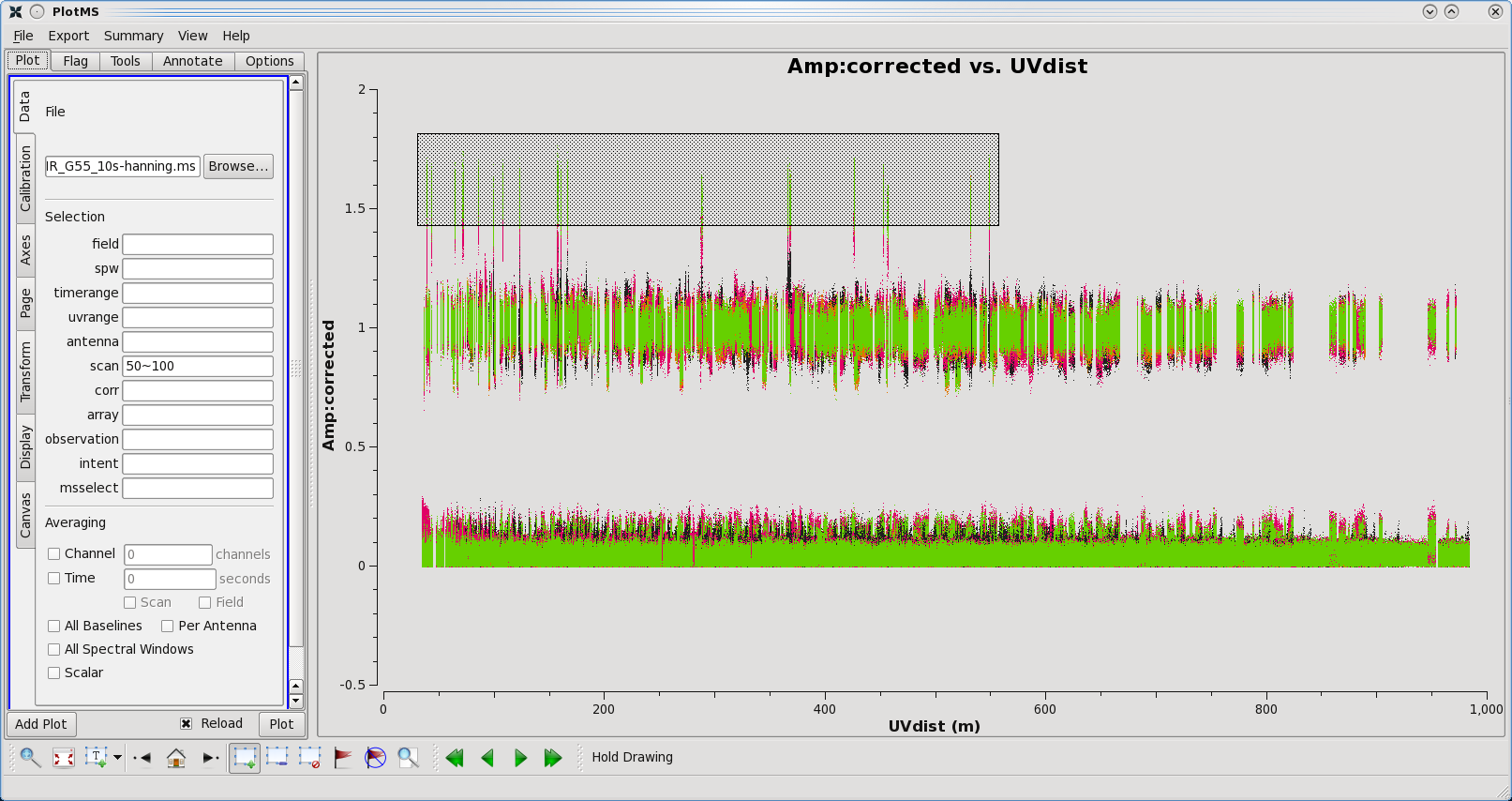
Let's again inspect amplitude vs uv-distance of our calibrated target data (Figure | Let's again inspect amplitude vs uv-distance of our calibrated target data (Figure 21) with plotms(). Instead of flagging the data interactively (the Flag button), it is recommended to run the [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_flagdata/about flagdata] task. While locating the bad data points in the previous section (<b>Mark Region</b> and then click <b>Locate</b> button), CASA logger provided us with information on the data points. Notice that they all belong to scans 88 and 87. The scans also involve a baseline shared with ea16. To flag these data run the flagdata() task: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 922: | Line 933: | ||
</source> | </source> | ||
To see the effect of this flagging, either | To see the effect of this flagging, either Reload the image (tick Reload and click Plot) if your plotms() session from the previous section is still open, or otherwise run the following command: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 930: | Line 941: | ||
</source> | </source> | ||
We can now see the image (Figure | We can now see the image (Figure 22) looks better without scans 87 and 88 for ea16. | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:Corr.Amp_vs_UVdist.png|400px|thumb|right|'''Figure | | [[Image:Corr.Amp_vs_UVdist.png|400px|thumb|right|'''Figure 21''' <br />Amplitude vs. UV-Distance plot for scans 50-100. Outliers have been highlighted to show interactive flagging through plotms.]] | ||
| [[Image:Corr.Amp_vs_UVdist.flagged.png|400px|thumb|right|'''Figure | | [[Image:Corr.Amp_vs_UVdist.flagged.png|400px|thumb|right|'''Figure 22''' <br /> Amplitude vs. UV-Distance plot for scans 50-100. Scans 87 and 88 have been flagged for ea16 via flagdata.]] | ||
|} | |} | ||
If you would like to perform these steps with [https://casa.nrao.edu/casadocs/casa-5.4.0/global-task-list/task_msview/about msview()], instead of plotms(), plot <b>time vs. baseline length</b>. Here baseline length is equivalent to the uv-distance. | |||
<!-- Now would be also a good time to play with the plotms parameters and look for more data to flag interactively. As a reminder, at the start of this guide we highlighted and located some of the worst sections with RFI. When reducing/flagging your data, it will prove helpful to review this information and revisit the most RFI affected spectral windows and channels.--> | |||
<!-- | |||
=== Amplitude vs. Baseline === | === Amplitude vs. Baseline === | ||
| Line 969: | Line 982: | ||
flagdata(vis='SNR_G55_10s-hanning.ms', antenna='ea02&ea08', spw='1', flagbackup=False) | flagdata(vis='SNR_G55_10s-hanning.ms', antenna='ea02&ea08', spw='1', flagbackup=False) | ||
</source> | </source> | ||
--> | |||
== Flagging Summary & Report == | == Flagging Summary & Report == | ||
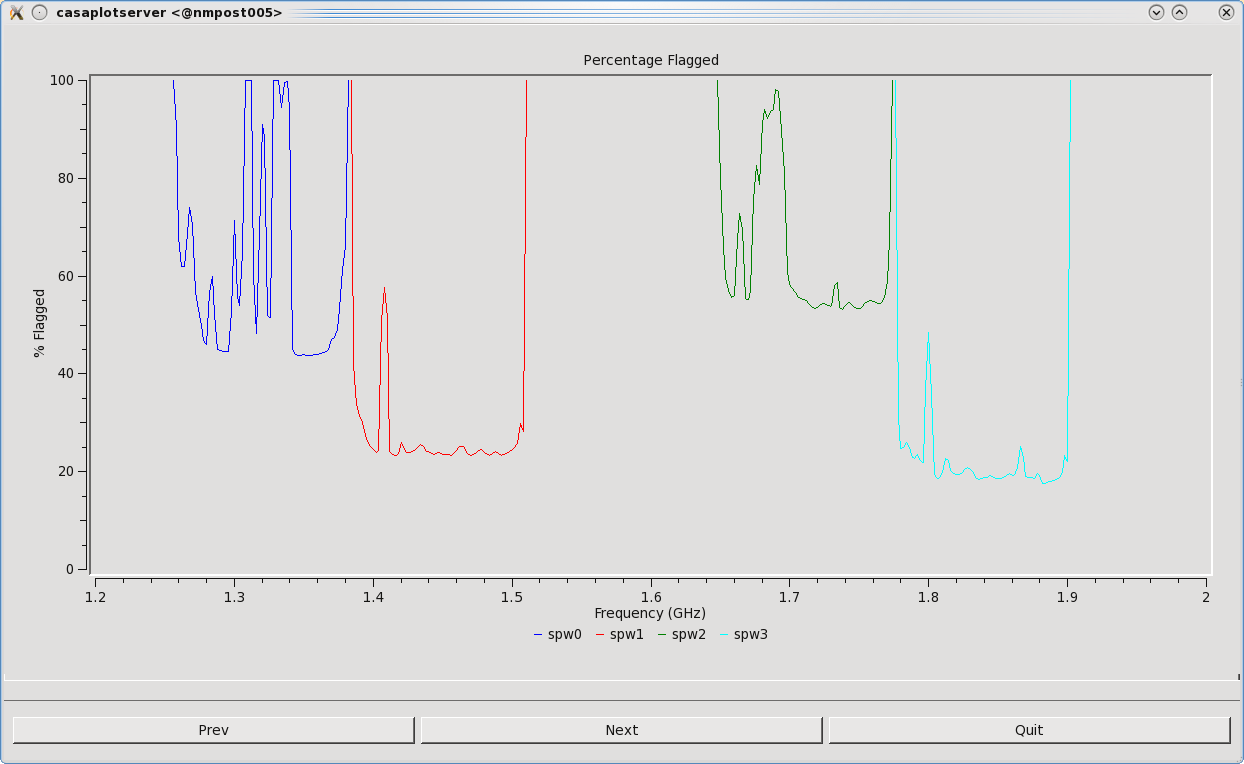
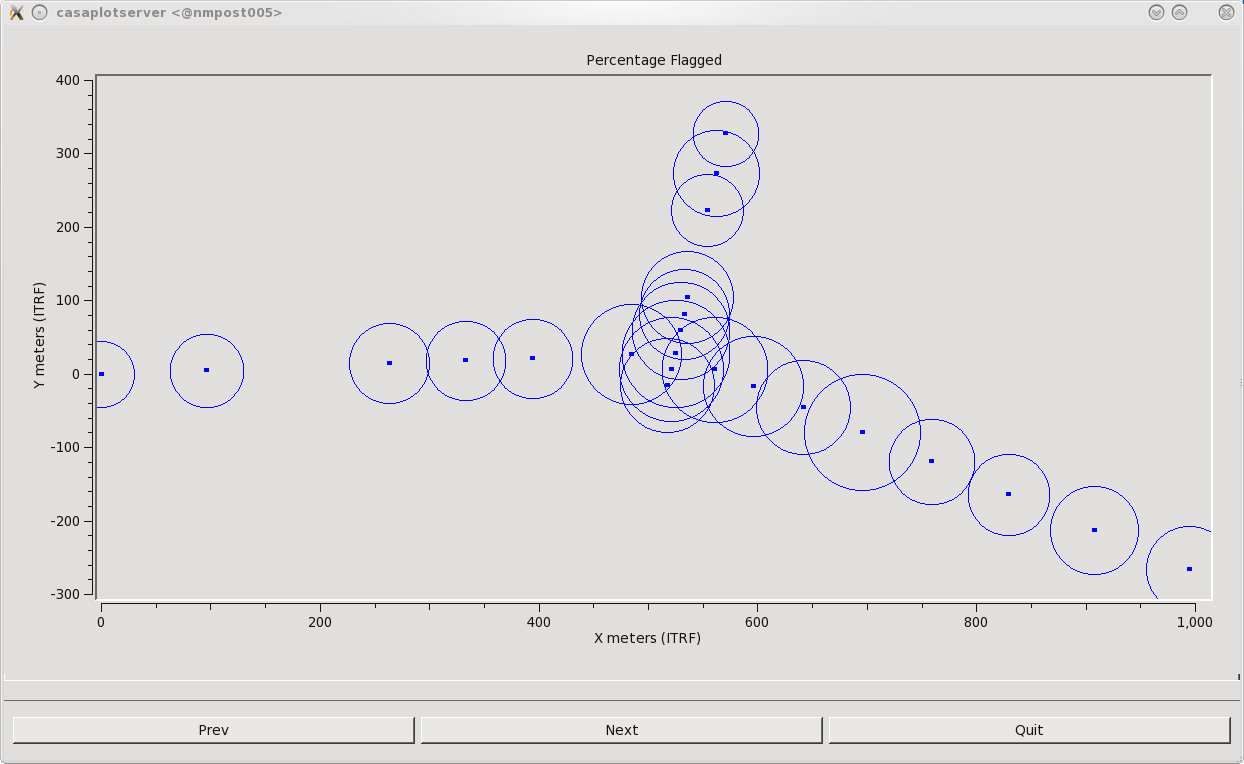
We will now run flagdata in summary mode to inspect how much total data has been flagged. We can also create a plot of the percentage of flagged data with respect to frequency by setting parameter ''display='report' ''(Figure | We will now run flagdata in summary mode to inspect how much total data has been flagged. We can also create a plot of the percentage of flagged data with respect to frequency by setting parameter ''display='report' ''(Figure 23). The report parameter will also create a plot of the antennas, with the circle size representing the percentage of flagged data per antenna (Figure 24). This can be viewed by clicking Next. One can also zoom in and out for a better view. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 1,004: | Line 1,015: | ||
{|style="margin: 0 auto;" | {|style="margin: 0 auto;" | ||
| [[Image:Percent.Flagged.Plot.png|400px|thumb|right|'''Figure | | [[Image:Percent.Flagged.Plot.png|400px|thumb|right|'''Figure 23''' <br /> Total percentage of flagged data vs. frequency, colorized by spectral window.]] | ||
| [[Image:Percent.Flagged.Ant.png|400px|thumb|right|'''Figure | | [[Image:Percent.Flagged.Ant.png|400px|thumb|right|'''Figure 24''' <br /> Total Percentage of data flagged from each antenna. A larger circle represents more flagged data for that antenna. ]] | ||
|} | |} | ||
| Line 1,031: | Line 1,042: | ||
--Edits: Jose Salcido (4.7.0, 2016/10/26) <br /> | --Edits: Jose Salcido (4.7.0, 2016/10/26) <br /> | ||
--Edits: Tony Perreault (4.7.0, 2016/10/26 <br /> | --Edits: Tony Perreault (4.7.0, 2016/10/26 <br /> | ||
--Edits: Anna Kapinska (5.4.0, 2018/ | --Edits: Anna Kapinska (5.4.0, 2018/11/30 <br /> | ||
--> | --> | ||
Latest revision as of 23:00, 30 November 2018
- This CASA guide is designed for CASA 5.4.0
- Printing Disclaimer: When printing, CASA commands in this guide may be cropped depending on your web browser text size and printer settings.
Overview
This CASA guide covers online data flagging, shadowing, zero-clipping, and quacking. It also covers auto-flagging RFI (Radio Frequency Interference) via TFCrop (Time-Frequency Crop) and rflag.
We will be utilizing data taken, with the Karl G. Jansky Very Large Array, of G055.7+3.4., which is a supernova remnant. The data were taken on August 23, 2010 in the first D-configuration for which the new wideband capabilities of the WIDAR (Wideband Interferometric Digital ARchitecture) correlator were available. The 8-hour long session included all available 1 GHz of bandwidth in L-band, from 1–2 GHz in frequency., although in this guide we will use dataset composed of only 4 spectral windows (128 MHz bandwidth each).
The guide will often reference the CASA documentation archives which are available online.
Obtaining the Data
A copy of the data can be downloaded from http://casa.nrao.edu/Data/EVLA/SNRG55/SNR_G55_10s.tar.gz (5.0 GB). These data are already in the CASA Measurement Set (MS) format and were prepared specifically for this guide.
The following explains how the data were processed to achieve the above MS file, as it has implications for later flagging steps.
Note that commands with a grey background do not need to be run in CASA.
The data were first loaded from the NRAO archive. On the archive search form, we specified Archive File ID to be AB1345_sb1800808_1.55431.004049953706 and selected SDM-BDF dataset (all files) as the data download option on the following page. Be aware that the full dataset is 170GB!
Once the SDM-BDF was downloaded, we created an MS with the task importasdm:
importasdm(asdm='AB1345_sb1800808_1.55431.004049953706', vis='SNR_G55.ms', process_flags=True, tbuff=1.5, applyflags=False,
ocorr_mode='co', savecmds=True, outfile='SNR_G55.ms.onlineflags.txt', flagbackup=False)
process_flags=True: This parameter controls the creation of online flags from the Flag.xml SDM table.
It will create online flags in the FLAG_CMD sub-table within the MS (more on this later).
tbuff=1.5: This parameter adds a time buffer padding to the flags in both directions to deal with timing mismatches.
This is important for VLA data taken before April 2011. This value should be set to 1.5x integration time.
This particular observing session had 1 second correlator integration time.
applyflags=False: We will apply these flags later in the tutorial.
ocorr_mode ='co': Only choosing to import the cross-only (co) correlations.
savecmds=True: Save the online-flag commands to an ASCII file.
outfile='SNR_G55.ms.onlineflags.txt': This will create a text file with a list of online flags that can be applied to the data.
The online flags within the file will include the time buffer requested in the tbuff parameter.
Mainly created for demonstration purposes.
The data were split from the original MS and time-averaged using the CASA task split.
split(vis='SNR_G55.ms', outputvis='SNR_G55_10s.ms', field='3~5', spw='4~5,7~8', timebin='10s',
antenna='!ea06,ea17,ea20,ea26', datacolumn='data', keepflags=False)
field='3~5': We only include the complex gain calibrator, flux density/bandpass calibrator, and the target source at this stage.
spw='4~5,7~8': Several spectral windows have been removed which were heavily impacted with RFI, which neither auto-flagging nor hand flagging could remedy. We are keeping spectral windows 4,5,7,8.
timebin='10s': Time-averaged to 10-seconds, to reduce size and processing time when running tasks.
antenna='!ea06,ea17,ea20,ea26': Several antennas have been removed, which at the time of the observations, did not have an L-Band receiver installed (please see the Identifying Problematic Antennas from the Operator Logs section). The exclamation point (!) before ea06 is called a negation operator. It will exclude all the antennas provided here, as long as they are separated by commas. For more on CASA syntax, please see this page on MS selection syntax.
Unpacking the Data
Once you've downloaded the MS file from the link above, unzip and untar the file from a terminal window (before starting CASA):
tar -xzvf SNR_G55_10s.tar.gz
This will create a directory called "SNR_G55_10s.ms" (5.5GB), which is the MS.
Starting CASA
Start CASA by typing casa on a terminal command line. If you have not used CASA before, some helpful tips are available on the Getting Started in CASA page.
This guide has been written for CASA version 5.4.0. Please confirm your version before proceeding by checking the message in the command line interface window or the CASA logger after startup.
For this tutorial, we will be running tasks using the task (parameter = value) syntax. All parameters not explicitly called by this manner will use their default values.
Preliminary Data Evaluation
As a first step, use listobs to have a look at the MS:
# In CASA
listobs(vis='SNR_G55_10s.ms', listfile='SNR_G55_10s.listobs')
- listfile: Creates a text document SNR_G55_10s.listobs with the summary of the data set.
The SNR_G55_10s.listobs text document can be opened with your favorite text viewer.
================================================================================
MeasurementSet Name: /lustre/aoc/sciops/akapinsk/CASAguides-5.4.0/flagging/SNR_G55_10s.ms MS Version 2
================================================================================
Observer: Dr. Sanjay Sanjay Bhatnagar Project: uid://evla/pdb/1072564
Observation: EVLA
Data records: 2732400 Total elapsed time = 26926 seconds
Observed from 23-Aug-2010/00:56:36.0 to 23-Aug-2010/08:25:22.0 (UTC)
ObservationID = 0 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows SpwIds Average Interval(s) ScanIntent
23-Aug-2010/00:56:36.0 - 00:58:06.0 14 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
00:58:06.0 - 00:59:36.0 15 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
00:59:36.0 - 01:01:05.0 16 0 J1925+2106 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [CALIBRATE_PHASE#UNSPECIFIED]
01:01:05.0 - 01:02:35.0 17 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
01:02:35.0 - 01:04:05.0 18 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
01:04:05.0 - 01:05:34.0 19 0 J1925+2106 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [CALIBRATE_PHASE#UNSPECIFIED]
01:05:34.0 - 01:07:04.0 20 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
01:07:04.0 - 01:08:34.0 21 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:08:34.0 - 01:10:04.0 22 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:10:04.0 - 01:11:34.0 23 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:11:34.0 - 01:13:03.0 24 1 G55.7+3.4 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [OBSERVE_TARGET#UNSPECIFIED]
01:13:03.0 - 01:14:33.0 25 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:14:33.0 - 01:16:03.0 26 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:16:03.0 - 01:17:33.0 27 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:17:33.0 - 01:19:02.0 28 1 G55.7+3.4 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [OBSERVE_TARGET#UNSPECIFIED]
01:19:02.0 - 01:20:32.0 29 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:20:32.0 - 01:22:02.0 30 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:22:02.0 - 01:23:32.0 31 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:23:32.0 - 01:25:01.0 32 1 G55.7+3.4 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [OBSERVE_TARGET#UNSPECIFIED]
01:25:01.0 - 01:26:31.0 33 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:26:31.0 - 01:28:01.0 34 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:28:01.0 - 01:29:31.0 35 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:29:31.0 - 01:31:00.0 36 1 G55.7+3.4 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [OBSERVE_TARGET#UNSPECIFIED]
01:31:00.0 - 01:32:30.0 37 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:32:30.0 - 01:34:00.0 38 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:34:00.0 - 01:35:30.0 39 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
01:35:30.0 - 01:36:59.0 40 1 G55.7+3.4 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [OBSERVE_TARGET#UNSPECIFIED]
01:36:59.0 - 01:38:29.0 41 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
01:38:29.0 - 01:39:59.0 42 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
01:39:59.0 - 01:41:29.0 43 0 J1925+2106 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_PHASE#UNSPECIFIED]
01:41:29.0 - 01:42:58.0 44 1 G55.7+3.4 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [OBSERVE_TARGET#UNSPECIFIED]
[....]
08:11:54.0 - 08:13:24.0 305 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
08:13:24.0 - 08:14:54.0 306 1 G55.7+3.4 9108 [0,1,2,3] [10, 10, 10, 10] [OBSERVE_TARGET#UNSPECIFIED]
08:14:54.0 - 08:16:24.0 307 2 0542+498=3C147 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
08:16:24.0 - 08:17:54.0 308 2 0542+498=3C147 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
08:17:54.0 - 08:19:23.0 309 2 0542+498=3C147 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
08:19:23.0 - 08:20:53.0 310 2 0542+498=3C147 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
08:20:53.0 - 08:22:23.0 311 2 0542+498=3C147 9108 [0,1,2,3] [10, 10, 10, 10] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
08:22:23.0 - 08:23:52.0 312 2 0542+498=3C147 9108 [0,1,2,3] [9.89, 9.89, 9.89, 9.89] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
08:23:52.0 - 08:25:22.0 313 2 0542+498=3C147 9108 [0,1,2,3] [10,10,10,10] [CALIBRATE_AMPLI#UNSPECIFIED,CALIBRATE_BANDPASS]
(nRows = Total number of rows per scan)
Fields: 3
ID Code Name RA Decl Epoch SrcId nRows
0 D J1925+2106 19:25:59.605371 +21.06.26.16218 J2000 0 391644
1 NONE G55.7+3.4 19:21:40.000000 +21.45.00.00000 J2000 1 2277000
2 N 0542+498=3C147 05:42:36.137916 +49.51.07.23356 J2000 2 63756
Spectral Windows: (4 unique spectral windows and 1 unique polarization setups)
SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) BBC Num Corrs
0 Subband:0 64 TOPO 1256.000 2000.000 128000.0 1319.0000 4 RR RL LR LL
1 Subband:2 64 TOPO 1384.000 2000.000 128000.0 1447.0000 4 RR RL LR LL
2 Subband:1 64 TOPO 1648.000 2000.000 128000.0 1711.0000 8 RR RL LR LL
3 Subband:0 64 TOPO 1776.000 2000.000 128000.0 1839.0000 8 RR RL LR LL
Sources: 12
ID Name SpwId RestFreq(MHz) SysVel(km/s)
0 J1925+2106 0 - -
0 J1925+2106 1 - -
0 J1925+2106 2 - -
0 J1925+2106 3 - -
1 G55.7+3.4 0 - -
1 G55.7+3.4 1 - -
1 G55.7+3.4 2 - -
1 G55.7+3.4 3 - -
2 0542+498=3C147 0 - -
2 0542+498=3C147 1 - -
2 0542+498=3C147 2 - -
2 0542+498=3C147 3 - -
Antennas: 23:
ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m)
East North Elevation x y z
0 ea01 W09 25.0 m -107.37.25.2 +33.53.51.0 -521.9416 -332.7766 -1.2001 -1601710.017000 -5042006.925200 3554602.355600
1 ea02 E02 25.0 m -107.37.04.4 +33.54.01.1 9.8240 -20.4293 -2.7806 -1601150.060300 -5042000.619800 3554860.729400
2 ea03 E09 25.0 m -107.36.45.1 +33.53.53.6 506.0564 -251.8670 -3.5825 -1600715.950800 -5042273.187000 3554668.184500
3 ea04 W01 25.0 m -107.37.05.9 +33.54.00.5 -27.3562 -41.3030 -2.7418 -1601189.030140 -5042000.493300 3554843.425700
4 ea05 W08 25.0 m -107.37.21.6 +33.53.53.0 -432.1167 -272.1478 -1.5054 -1601614.091000 -5042001.652900 3554652.509300
5 ea07 E05 25.0 m -107.36.58.4 +33.53.58.8 164.9788 -92.8032 -2.5268 -1601014.462000 -5042086.252000 3554800.799800
6 ea08 N01 25.0 m -107.37.06.0 +33.54.01.8 -30.8810 -1.4664 -2.8597 -1601185.634945 -5041978.156586 3554876.424700
7 ea09 E06 25.0 m -107.36.55.6 +33.53.57.7 236.9058 -126.3369 -2.4443 -1600951.588000 -5042125.911000 3554773.012300
8 ea10 N03 25.0 m -107.37.06.3 +33.54.04.8 -39.0773 93.0192 -3.3330 -1601177.376760 -5041925.073200 3554954.584100
9 ea11 E04 25.0 m -107.37.00.8 +33.53.59.7 102.8054 -63.7682 -2.6414 -1601068.790300 -5042051.910200 3554824.835300
10 ea12 E08 25.0 m -107.36.48.9 +33.53.55.1 407.8285 -206.0065 -3.2272 -1600801.926000 -5042219.366500 3554706.448200
11 ea13 N07 25.0 m -107.37.07.2 +33.54.12.9 -61.1037 344.2331 -4.6138 -1601155.635800 -5041783.843800 3555162.374100
12 ea15 W06 25.0 m -107.37.15.6 +33.53.56.4 -275.8288 -166.7451 -2.0590 -1601447.198000 -5041992.502500 3554739.687600
13 ea16 W02 25.0 m -107.37.07.5 +33.54.00.9 -67.9687 -26.5614 -2.7175 -1601225.255200 -5041980.383590 3554855.675000
14 ea18 N09 25.0 m -107.37.07.8 +33.54.19.0 -77.4346 530.6273 -5.5859 -1601139.485100 -5041679.036800 3555316.533200
15 ea19 W04 25.0 m -107.37.10.8 +33.53.59.1 -152.8599 -83.8054 -2.4614 -1601315.893000 -5041985.320170 3554808.304600
16 ea21 E01 25.0 m -107.37.05.7 +33.53.59.2 -23.8638 -81.1510 -2.5851 -1601192.467800 -5042022.856800 3554810.438800
17 ea22 N04 25.0 m -107.37.06.5 +33.54.06.1 -42.6239 132.8436 -3.5494 -1601173.979400 -5041902.657700 3554987.517500
18 ea23 E07 25.0 m -107.36.52.4 +33.53.56.5 318.0509 -164.1850 -2.6957 -1600880.571400 -5042170.388000 3554741.457400
19 ea24 W05 25.0 m -107.37.13.0 +33.53.57.8 -210.0959 -122.3887 -2.2577 -1601377.009500 -5041988.665500 3554776.393400
20 ea25 N02 25.0 m -107.37.06.2 +33.54.03.5 -35.6245 53.1806 -3.1345 -1601180.861480 -5041947.453400 3554921.628700
21 ea27 E03 25.0 m -107.37.02.8 +33.54.00.5 50.6641 -39.4835 -2.7273 -1601114.365500 -5042023.151800 3554844.944000
22 ea28 N08 25.0 m -107.37.07.5 +33.54.15.8 -68.9057 433.1889 -5.0602 -1601147.940400 -5041733.837000 3555235.956000
- J1925+2106, field ID 0: Phase (complex gain (amplitude and phase)) calibrator;
- G55.7+3.4, field ID 1: The supernova remnant;
- 0542+498=3C147, field ID 2: Flux density/bandpass calibrator.
Antenna plot showing the configuration during the observing session.
We can also see that these sources have associated scan intents that indicate their function in the observations. Note that you can select sources based on their intents in certain CASA tasks. The various scan intents in this data set are:
- CALIBRATE_PHASE indicates that this is a scan to be used for complex gain (amplitude and phase) calibration;
- OBSERVE_TARGET indicates that this is the science target;
- CALIBRATE_AMPLI indicates that this is to be used for flux density calibration, and;
- CALIBRATE_BANDPASS indicates that these scans are to be used for bandpass calibration.
Note that more recent data sets may have different scan intents assigned to calibration scans.
It's important to be aware that the antennas have both a name and an ID associated with them. For example antenna ID 15 is named ea19 (the ea stemming from the Expanded VLA project). When specifying an antenna within a task parameter, we will primarily reference them by their ea name.
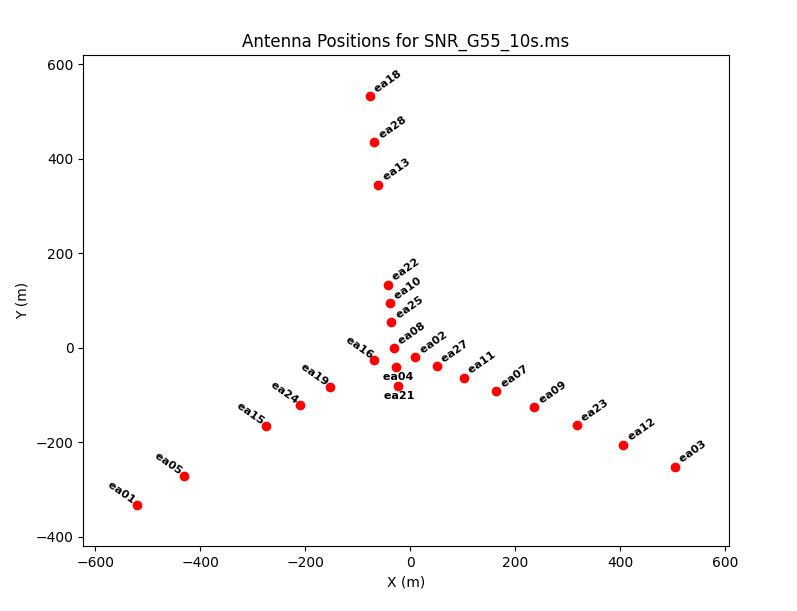
We can see the antenna configuration for this observing session by using plotants.
# In CASA
plotants(vis='SNR_G55_10s.ms', figfile='SNR_G55_10s.plotants.png')
Figure 1 the antenna configuration for our observations; antennas ea01, ea03, and ea18 were on the extreme ends of the west, east, and north arms, respectively.
Identifying RFI with Plotms()
Plotms image of supernova remnant data, showing strong RFI through the observation (x axis) and affecting a number of spectral windows (data are colorised by spw).
The data can be inspected with the plotms task.
It is worth first having a look at the data, separately per each field, in amplitude vs. frequency (to identify narrow-band RFI) and amplitude vs. time (intermittent RFI).
To start with, let's look at the supernova remnant observations (field='1') for all scans (default) and all antennas (default):
# In CASA
plotms(vis='SNR_G55_10s.ms', field='1', xaxis='time', yaxis='amp', coloraxis='spw')
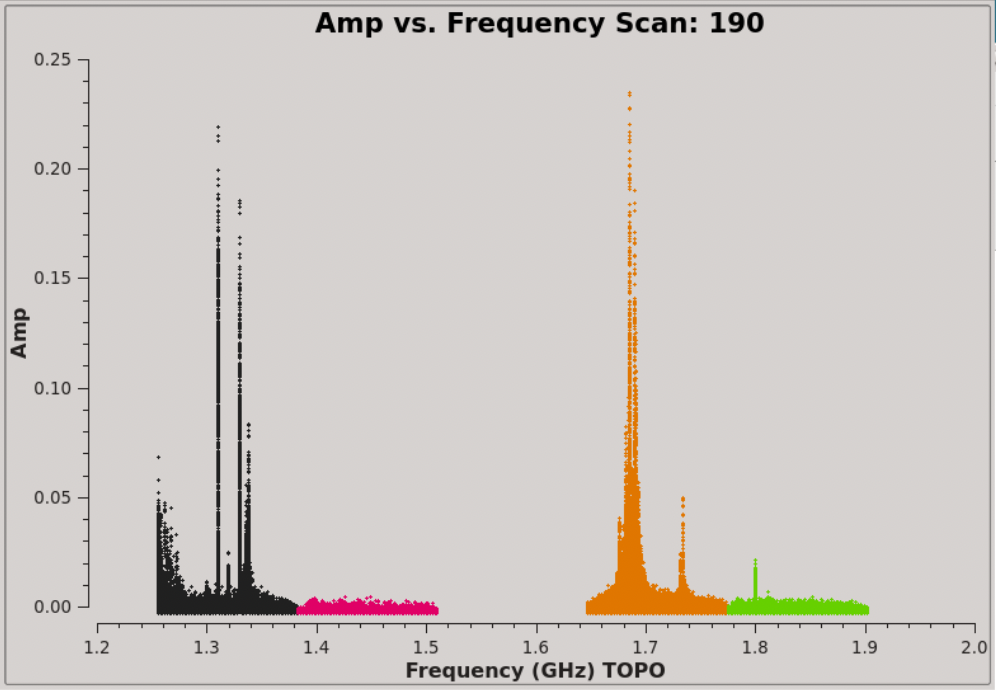
We can immediately see that there is plenty of RFI throughout the observations, affecting a number of spectral windows (data are colorised by spectral window; Figure 2). The RFI is discerned by the large spikes in amplitude.
To narrow down where the RFI occurs let's now have a look at the data in amplitude vs frequency, plotted separately per each scan:
# In CASA
plotms(vis='SNR_G55_10s.ms', scan='30,75,120,165,190,235,303', antenna='ea24', xaxis='freq',
yaxis='amp', coloraxis='spw', iteraxis='scan')
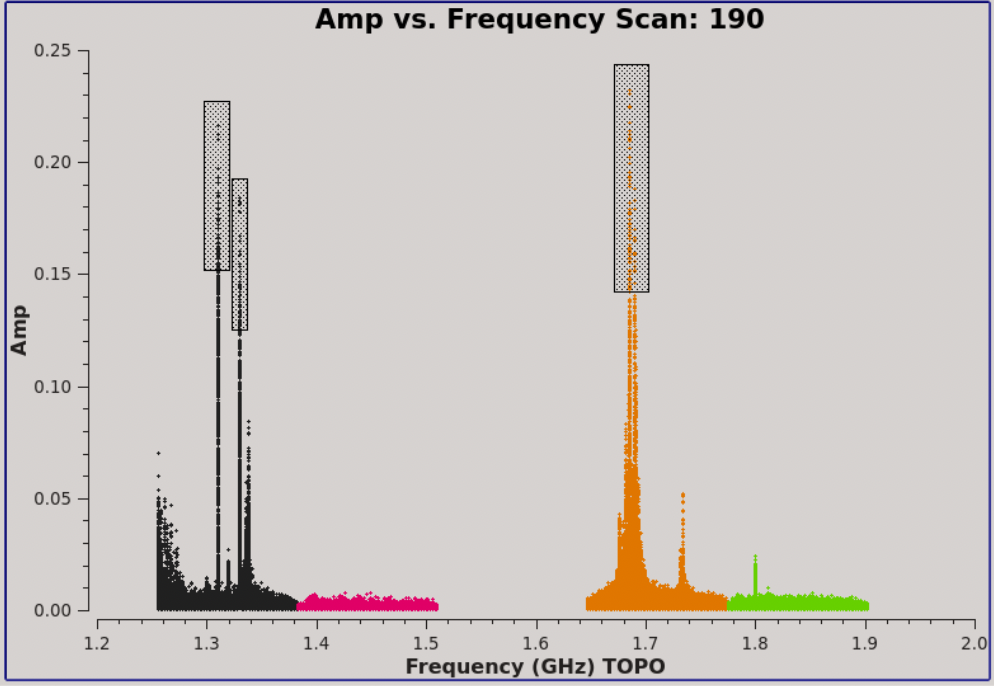
Plotms image of scan 190 showing strong RFI spikes in amplitude for several spectral windows.
- coloraxis='spw': Parameter indicates that a different color will be assigned to each spectral window.
- antenna='ea24' : Here we chose only information for antenna ea24.
- iteraxis='scan': Parameter tells plotms to iterate over scans, that is to display a new plot for each scan.
The command above will set the plotms for iteration, but if you want to set this option from within plotms, go onto the Plot tab (horizontal), and then Page tab (vertical). You will now find Iteration options. Set Axis to the parameter you want to iterate through. Iterate by clicking on the green arrow buttons on the toolbar within the plotms.
Progressing through to scan 190 , we can see there is significant time and frequency variable RFI present in the observing session. To determine which spectral windows are affected, click on the Mark Regions tool at the bottom of the plotms GUI (the open box with a green plus sign) and use the mouse to select a few of the highest-amplitude points in each of the spectral windows (Figure 3). Click on the Locate button (magnifying glass on white background). Information about the selected areas should now display in the logger window. For example:
INFO PlotMS Frequency in [1.30517 1.31368] or [1.32584 1.33313] or [1.68146 1.69544], Amp in [0.132051 0.223077] or [0.124359 0.188034] or [0.139316 0.237179]: INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea01@W09 & ea24@W05[0&19] Spw=0 Chan=27 Freq=1.31 Corr=LR X=1.31 Y=0.139029 (110/144/110) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea03@E09 & ea24@W05[2&19] Spw=0 Chan=27 Freq=1.31 Corr=LL X=1.31 Y=0.145391 (623/144/623) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea04@W01 & ea24@W05[3&19] Spw=0 Chan=27 Freq=1.31 Corr=LL X=1.31 Y=0.166033 (879/144/879) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea04@W01 & ea24@W05[3&19] Spw=0 Chan=37 Freq=1.33 Corr=LR X=1.33 Y=0.131179 (918/144/918) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea07@E05 & ea24@W05[5&19] Spw=0 Chan=27 Freq=1.31 Corr=RL X=1.31 Y=0.14596 (1389/144/1389) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea07@E05 & ea24@W05[5&19] Spw=0 Chan=27 Freq=1.31 Corr=LL X=1.31 Y=0.142578 (1391/144/1391) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea09@E06 & ea24@W05[7&19] Spw=0 Chan=27 Freq=1.31 Corr=RR X=1.31 Y=0.141398 (1900/144/1900) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea12@E08 & ea24@W05[10&19] Spw=0 Chan=37 Freq=1.33 Corr=LR X=1.33 Y=0.133163 (2710/144/2710) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea13@N07 & ea24@W05[11&19] Spw=0 Chan=37 Freq=1.33 Corr=RR X=1.33 Y=0.129079 (2964/144/2964) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea15@W06 & ea24@W05[12&19] Spw=0 Chan=27 Freq=1.31 Corr=RR X=1.31 Y=0.135901 (3180/144/3180) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea16@W02 & ea24@W05[13&19] Spw=0 Chan=27 Freq=1.31 Corr=RR X=1.31 Y=0.17187 (3436/144/3436) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea16@W02 & ea24@W05[13&19] Spw=0 Chan=27 Freq=1.31 Corr=LR X=1.31 Y=0.155113 (3438/144/3438) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea16@W02 & ea24@W05[13&19] Spw=0 Chan=27 Freq=1.31 Corr=LL X=1.31 Y=0.142598 (3439/144/3439) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea19@W04 & ea24@W05[15&19] Spw=0 Chan=27 Freq=1.31 Corr=LR X=1.31 Y=0.154241 (3950/144/3950) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea19@W04 & ea24@W05[15&19] Spw=0 Chan=27 Freq=1.31 Corr=LL X=1.31 Y=0.178607 (3951/144/3951) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea19@W04 & ea24@W05[15&19] Spw=0 Chan=37 Freq=1.33 Corr=RR X=1.33 Y=0.126879 (3988/144/3988) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea21@E01 & ea24@W05[16&19] Spw=0 Chan=27 Freq=1.31 Corr=RL X=1.31 Y=0.144868 (4205/144/4205) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea21@E01 & ea24@W05[16&19] Spw=0 Chan=27 Freq=1.31 Corr=LL X=1.31 Y=0.158044 (4207/144/4207) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:19:58.0 BL=ea22@N04 & ea24@W05[17&19] Spw=0 Chan=37 Freq=1.33 Corr=RR X=1.33 Y=0.18105 (4500/144/4500) <snip> INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:08.0 BL=ea22@N04 & ea24@W05[17&19] Spw=2 Chan=19 Freq=1.686 Corr=RR X=1.686 Y=0.195303(10060/169/4428) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:08.0 BL=ea22@N04 & ea24@W05[17&19] Spw=2 Chan=19 Freq=1.686 Corr=RL X=1.686 Y=0.19185 (10061/169/4429) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:08.0 BL=ea22@N04 & ea24@W05[17&19] Spw=2 Chan=21 Freq=1.69 Corr=RR X=1.69 Y=0.169598 (10068/169/4436) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea08@N01 & ea24@W05[6&19] Spw=2 Chan=19 Freq=1.686 Corr=LR X=1.686 Y=0.157531(7246/170/1614) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea10@N03 & ea24@W05[8&19] Spw=2 Chan=19 Freq=1.686 Corr=LL X=1.686 Y=0.15168 (7759/170/2127) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea15@W06 & ea24@W05[12&19] Spw=2 Chan=19 Freq=1.686 Corr=LR X=1.686 Y=0.202071(8782/170/3150) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea18@N09 & ea24@W05[14&19] Spw=2 Chan=21 Freq=1.69 Corr=RR X=1.69 Y=0.159526 (9300/170/3668) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea19@W04 & ea24@W05[15&19] Spw=2 Chan=19 Freq=1.686 Corr=RR X=1.686 Y=0.192973(9548/170/3916) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea19@W04 & ea24@W05[15&19] Spw=2 Chan=19 Freq=1.686 Corr=LR X=1.686 Y=0.155768(9550/170/3918) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea19@W04 & ea24@W05[15&19] Spw=2 Chan=19 Freq=1.686 Corr=LL X=1.686 Y=0.147995(9551/170/3919) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea22@N04 & ea24@W05[17&19] Spw=2 Chan=19 Freq=1.686 Corr=RR X=1.686 Y=0.199225(10060/170/4428) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea22@N04 & ea24@W05[17&19] Spw=2 Chan=19 Freq=1.686 Corr=RL X=1.686 Y=0.193489(10061/170/4429) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea22@N04 & ea24@W05[17&19] Spw=2 Chan=21 Freq=1.69 Corr=RR X=1.69 Y=0.179036 (10068/170/4436) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea24@W05 & ea25@N02[19&20] Spw=2 Chan=19 Freq=1.686 Corr=RL X=1.686 Y=0.156535(10573/170/4941) INFO PlotMS Scan=190 Field=G55.7+3.4[1] Time=2010/08/23/05:21:17.5 BL=ea24@W05 & ea25@N02[19&20] Spw=2 Chan=21 Freq=1.69 Corr=RR X=1.69 Y=0.140293 (10580/170/4948) INFO PlotMS Found 287 points (287 unflagged) among 202752 in 0.01s.
Reviewing the log output, we see that spw 0 and 2 are affected the most by RFI. We also get the corresponding frequency where the RFI is present, which appears to be 1.31 and 1.33 GHz for spectral window 0, and 1.686 GHz for spectral window 2. This information is important, especially when flagging data interactively as we will be doing towards the end of this guide.
The VLA has obtained RFI sweeps over its full frequency range. The results are available on the Radio Frequency Interference website. Most of the RFI features can be identified as radar, communications, satellites, airplanes, or birdies (i.e., RFI generated by the VLA electronics itself). For our observations, we inspect the L-band specific RFI plots website and find that the 1310 and 1330 MHz features are due to FAA ASR radars, while the one at 1686 MHz is most likely due to a GOES weather satellite. We can compare the plots of the FAA ASR radar from the website (Figure 4a), with that of spw 0 (Figure 4b), and see that the spikes are practically identical and fall within the same frequencies.
Identifying RFI with MSview()
Similar to using plotms() for the ms file editing, one can also use the msview() task.
MSview() displays data in a 3D fashion, where the intensity of the plotted data points represents amplitude, phase, or their derivatives, while x and y axes of the display can be set to any of the following: time, baseline, channel, correlation or spectral window. For the introduction to the msview() tool see its dedicated guide (note that msview() can also be invoked as viewer(), as it is done in the linked guide).
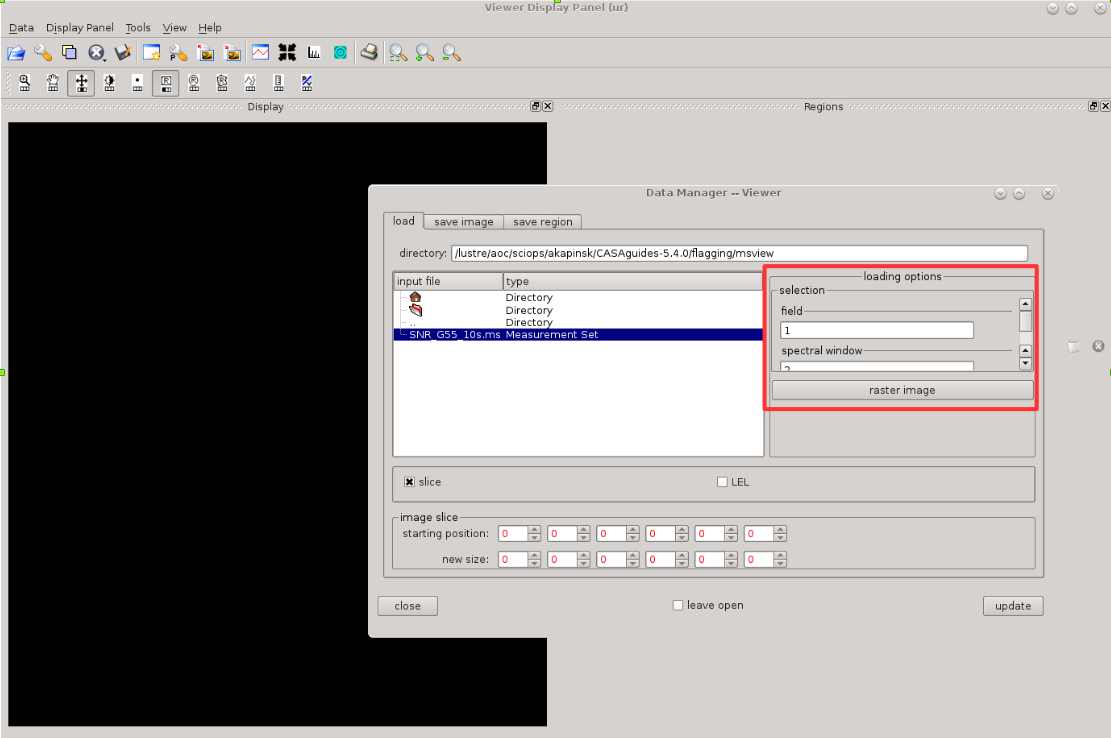
Loading data into msview(). The red box marks where data preselection can be made.
Note: msview() will not handle large data volumes very well, and it may crash if your ms file is too large. If that happens try preselecting a fraction of your data for viewing. Also, sometimes restarting CASA may help.
Since the task does not allow for much data selection in the command line, open msview() tool first, and load and select the data from its graphical interface:
# In CASA
msview()
Once the tool opens, it will prompt you for data upload. At the same time as selecting the ms file to open, choose which field, spw etc you would like to load from that ms file as shown in Figure 5, and hit Raster Image button. Only the preselected data will be available in the viewer, and further selection for viewing can be done via Data Display Options window (wrench button, within msview()) under its display axes tab. Note that the ms and visibility selection tab will inform you about the overall shape of your data, even if you have not loaded the full ms file.
To identify RFI in msview(), one can look at the data in multiple ways. For example, let's plot time vs. channel for a single spectral window. We can set these selections in the Data Display Options (click on the Wrench button) as shown in Figure 6. The selection window will appear (highlighted with red box in Figure 6). We plotted here spw=2, which corresponds to the orange-coloured spw from Figure 3. The blue coloured pixels are fairly well behaved data, while the green and red pixels are the data affected by the RFI. We can narrow down where that RFI occurs by hovering cursor over the RFI areas (white box/magenta circle in Figure 6) and details on affected baselines, scans, etc will be listed in the Cursors window (highlighted with magenta box in Figure 6). Note that msview refers to the antennas via their ID numbers and not the ea names (compare with the listobs output and commentary in section Preliminary Data Evaluation). Also, note that here only RR polarisation is plotted, and you will need to check also LL (as well as RL and LR if you have and use them in further data processing).
One can also have a quick look at which spectral windows most RFI occurs, and when. For example, let's plot channel vs. spectral window and iterate over time Figure 7). You can choose iteration by selecting animation axis under the display axes tab within the Data Display Options (marked with red box in Figure 7). To inspect how the RFI changes with time use the iteration buttons within the Animators panel (marked with yellow box in Figure 7). Again, the blue colour marks the well behaved data here; you can change the colour scheme in the Data Display Option window.
Identifying Problematic Antennas from the Operator Logs
We first check the operator log for the observing session to see if there were any issues noted during the run that need to be addressed. The logs are available from the VLA operator log website. Here is the link for the observing log file for our observing session.
The log has various pieces of information including the start/end times for the observing session, frequency bands used, weather, baseline information for recently moved antennas, and any outages or issues that may have been encountered. We see that antenna ea07 may need position corrections (see one of our calibration tutorials, IRC+10216, on how to fix this), and several antennas (ea06, ea17, ea20, and ea26) are missing an L-Band receiver. Antenna 07 position corrections will need to be applied during data calibration and will not be covered in this tutorial. Additionally, the antennas with missing receivers were already removed from the dataset used here (as explained in the above section Obtaining the Data), so we can continue with online flagging.
Online Flags
At the time of importing from the SDM-BDF raw data to a MS, we chose to process the online flags from the Flags.xml file to the FLAG_CMD sub-table within the MS. We also created the SNR_G55.ms.onlineflags.txt file, a plain text document that includes the list of online flags. Note that this step is not necessary if you choose to apply online flags at the time of the data download from the archive.
The Flags.xml file holds information of flags created during the observing session, such as subreflector issues and antennas not being on source. We will now apply these online flags to the data by employing flagcmd, but first, let's create a plot of the flags to get an idea of what will be flagged.
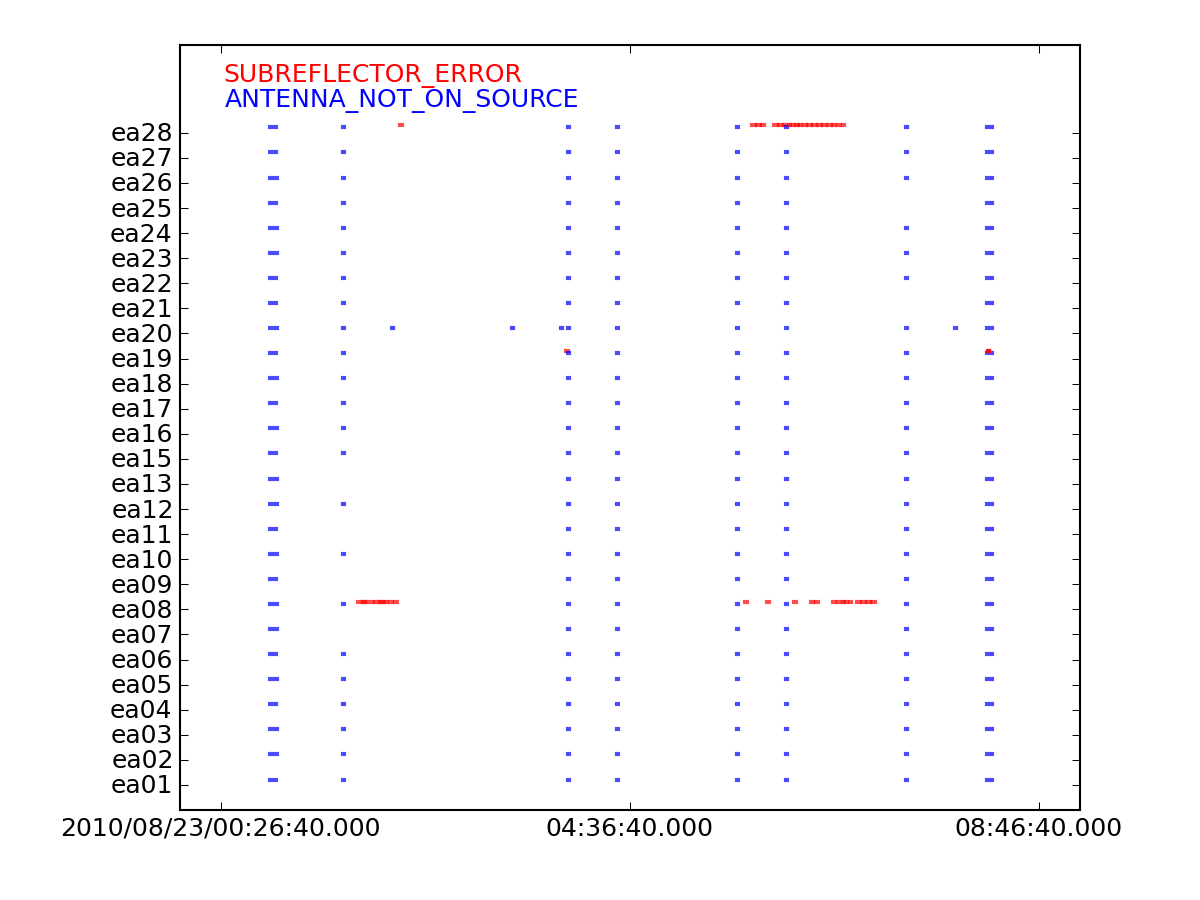
# In CASA
flagcmd(vis='SNR_G55_10s.ms', inpmode='table', reason='any', action='plot', plotfile='flaggingreason_vs_time.png')
Open the flaggingreason_vs_time.png with your favorite image viewing program. Figure 8 shows several instances of online flagging. Most notably, ea28 and ea08 had some subreflector issues throughout the observing session. Online flags are instances of possible missing data, including:
- ANTENNA_NOT_ON_SOURCE
The VLA antennas have slewing speeds of 40 degrees per minute in azimuth and 20 degrees per minute in elevation. Some antennas are slower than others and may take a few more seconds to reach the next source. The antennas can also take a few seconds to settle down due to small oscillations after having slewed.
- SUBREFLECTOR_ERROR
The Focus Rotation Mount (FRM), located at the apex of the antenna, is responsible for focusing the incoming radio signal to the corresponding receiver. At times there can be issues with the focus and/or rotation axes.
Now that we've plotted the online flags, we will apply them to the MS.
# In CASA
flagcmd(vis='SNR_G55_10s.ms', inpmode='table', reason='any', action='apply', flagbackup=False)
- inpmode='table': Will use the online-flags imported to the FLAG_CMD sub-table from the Flags.xml file. This was done during the importing of our data with importasdm. Note that the FLAG_CMD sub-table already includes a 1.5 second time buffer, as was requested during the importing of the data (parameter tbuff=1.5).
The CASA logger should report the progress as the task applies these flags in segments. Once finished, it will report the percentage of flagged data. The terminal window will display warnings about the four missing antennas; this is expected as we had previously removed the antennas during the split task.
Alternatively, we could have applied the online flags from the text file (SNR_G55.ms.onlineflags.txt) created during the importing of the data. The one caveat when it comes to applying flags from a list is not being able to un-apply them (setting parameter action='unapply' within flagcmd). Transferring the flags to the FLAGS_CMD table before applying them does allow for the use of the un-apply feature. For convenience, we provide the command for applying online flags from a list (don't run the following command in CASA):
flagcmd(vis='SNR_G55_10s.ms', inpmode='list', inpfile='SNR_G55.ms.onlineflags.txt', reason='any', action='apply', flagbackup=False)
Shadowed Antennas
The VLA D-configuration is the most compact; there may be instances where one antenna blocks, or shadows another. For more on shadowing, please refer to the antenna shadowing section in the Guide to Observing with the VLA. Generally, observing sources at 40 degrees elevation and higher will result in less shadowing. We will create a plot of elevation vs. time by using the plotms task and note the elevation of the radio sources we observed.
# In CASA
plotms(vis='SNR_G55_10s.ms', xaxis='time', yaxis='elevation', antenna='0&1;2&3', spw='*:31',
coloraxis='field', title='Elevation vs. Time', plotfile='Elevation_vs_Time.png')
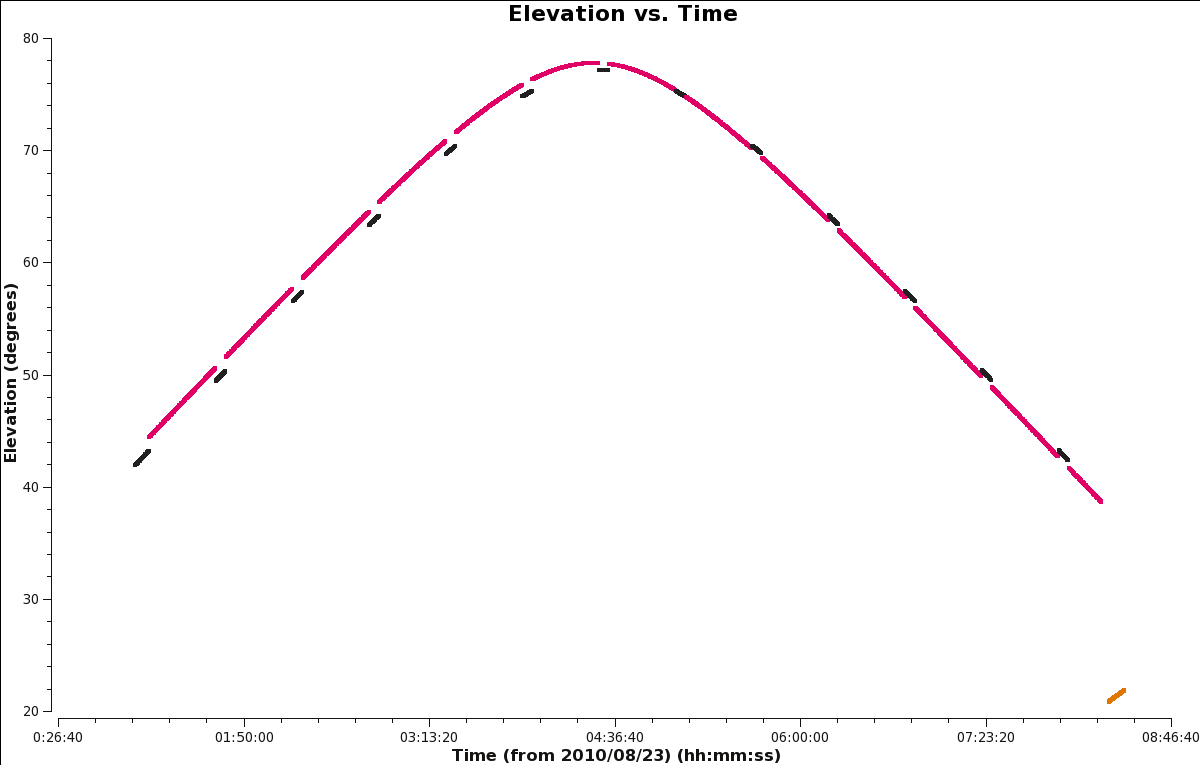
Figure 9 shows that most of the sources were at or above 40 degrees in elevation; accordingly, we expect a minimal amount of shadowing. The flux density / bandpass calibrator 3C147 was observed towards the end of the session, which could result in some shadowing of antennas due to its low elevation of about 20–22 degrees.
Task flagdata can determine and flag shadowed antennas by their location and observing direction:
# In CASA
flagdata(vis='SNR_G55_10s.ms', mode='shadow', tolerance=0.0, flagbackup=False)
- mode='shadow': has a subparameter tolerance=0.0 that controls how many meters of shadowing overlap is allowed
The logger will report on the percentage of flagged data due to shadowing. As expected in this observing session, there does not appear to be much data affected by shadowing (approx. 0.4%).
Zero-Amplitude Data
In addition to shadowing, there may be times during which the correlator writes out pure zero-valued data. In order to remove this bad data, we run flagdata to remove any pure zeroes:
# In CASA
flagdata(vis='SNR_G55_10s.ms', mode='clip', clipzeros=True, flagbackup=False)
- mode='clip': is used to flag all values below a given threshold. With clipzeros=True this mode will flag exact zero values that are sometimes being produced by the VLA correlator.
Inspecting the logger output generated by flagdata shows that there is a small quantity of zero-valued data present in this MS.
Quacking
Now we utilize the flagdata task one more time to run quacking mode.
It's common for the array to settle down at the start of a scan. Quacking is used to remove data at scan boundaries, the same edit can be applied to all scans for all baselines.
# In CASA
flagdata(vis='SNR_G55_10s.ms', mode='quack', quackinterval=5.0, quackmode='beg', flagbackup=False)
- quackmode='beg': Data from the start of each scan will be flagged.
- quackinterval=5.0: Flag the first 5 seconds of every scan.
Backup Data - Flagmanager
Flags can be backed up in a file ms.flagversions that is related to the MS. For our MS, the related flag backups are stored in SNR_G55_10s.ms.flagversions.
Note: flags that are applied to the data are contained in the MS; ms.flagversions only contains backup flags that can be restored if required.
Most flagging tasks, like flagdata, offer a flagbackup parameter that controls whether or not the current flags are being backed up in ms.flagversions before new (additional) flags will be applied. Backup flags can also be manually saved to or restored from ms.flagversions using the flagmanager task.
Now that we've applied online flags, clipped zero amplitude data, removed shadowed data, and quacked the data, we will create a backup of the flags by setting the parameter versionname='after_online_flagging' :
# In CASA
flagmanager(vis='SNR_G55_10s.ms', mode='save', versionname='after_online_flagging')
The version we just created is saved inside the SNR_G55_10s.ms.flagversions directory. From here onward, if we make a mistake, we can always revert back to this flag version of the MS by setting parameter mode='restore' and providing the version name we want to revert back to, such as:
flagmanager(vis='SNR_G55_10s.ms', mode='restore', versionname='after_online_flagging')
The flagmanager() can also be run on calibration tables. In this instance just specify the table name as the vis parameter and run is as above.
Benefit of Hanning Smoothing
Strong RFI sources can give rise to the Gibbs phenomenon. This is seen by ringing: a zig-zag pattern across the channels that neighbor the strong, usually narrow, RFI. To remedy this ringing across the frequency channels, we employ the Hanningsmooth algorithm via the hanningsmooth task (an implementation based on mstransform). Hanning-smoothing applies a triangle kernel across the pattern which diminishes the ringing, reducing the number of channels that may look bad and get flagged. Note that this smoothing procedure will also decrease the spectral resolution by a factor of two.
The task requires the creation of a new MS, which we will call SNR_G55_10s-hanning.ms.
Note: As Hanning-smoothing will remove amplitude spikes, it is not recommended for spectral analysis related science (e.g. strong narrow maser lines).
Let's create before (Figure 10a) and after images (Figure 10b) with the plotms task to see the effect.
# In CASA
plotms(vis='SNR_G55_10s.ms', scan='190', antenna='ea24', spw='0~2',
xaxis='freq', yaxis='amp', coloraxis='spw', title='Before Hanning',
correlation='RR,LL', plotrange=[1.2,1.8,-0.01,0.25],
plotfile='amp_v_freq.beforeHanning.png')
# In CASA
hanningsmooth(vis='SNR_G55_10s.ms', outputvis='SNR_G55_10s-hanning.ms', datacolumn='data')
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='190', antenna='ea24', spw='0~2',
xaxis='freq', yaxis='amp', coloraxis='spw', title='After Hanning',
correlation='RR,LL', plotrange=[1.2,1.8,-0.01,0.25],
plotfile='amp_v_freq.afterHanning.png')
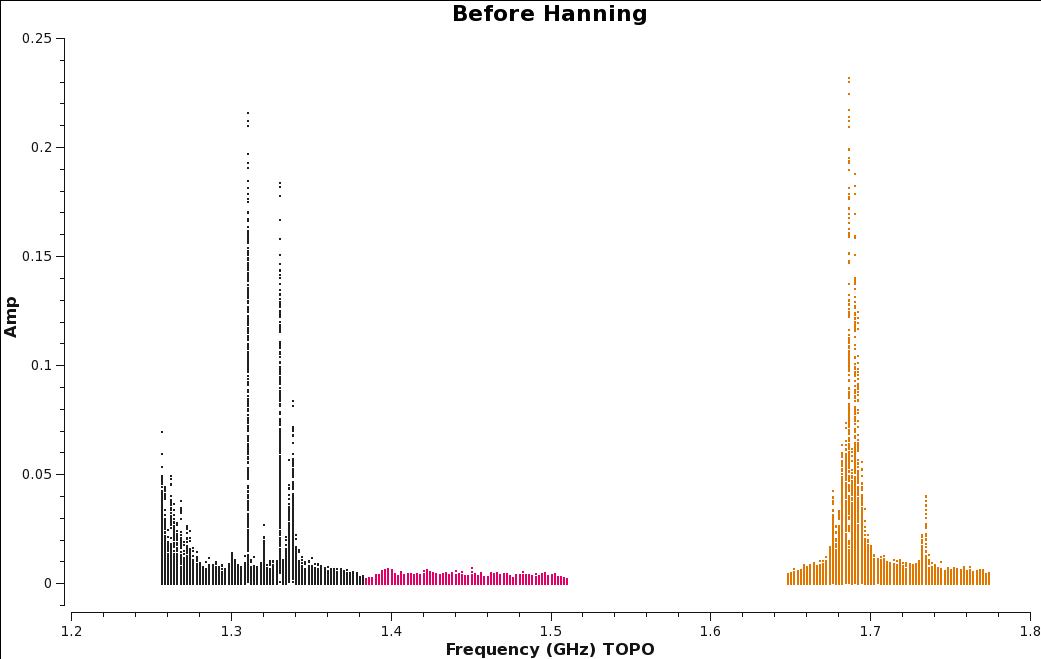 Plot of amplitude vs. frequency before Hanning smoothing data. |
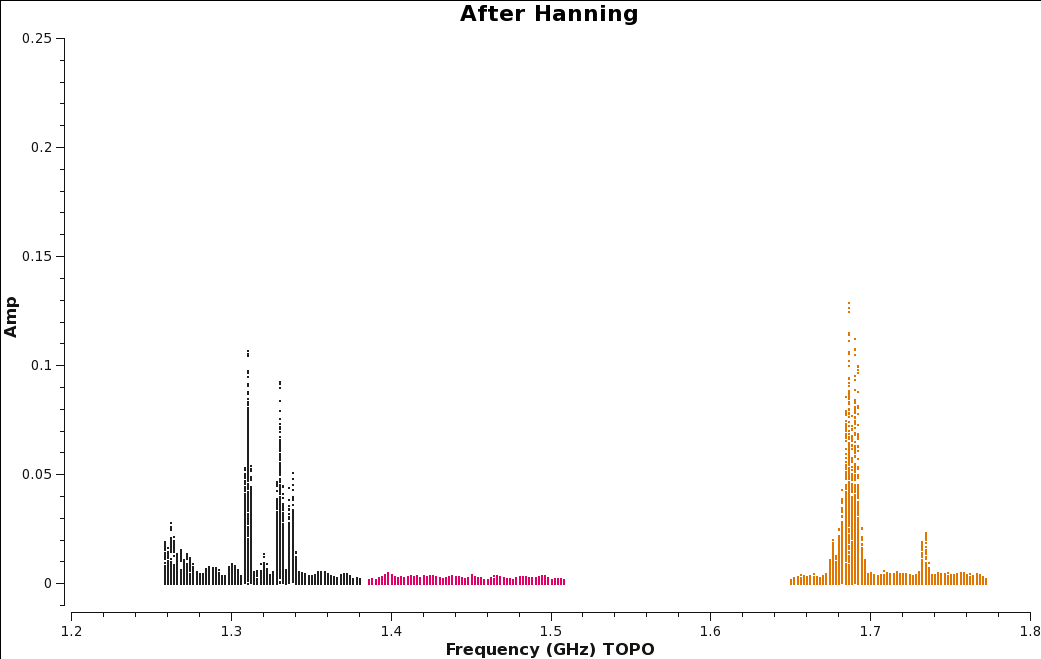 Plot of amplitude vs. frequency after Hanning smoothing data. |
Figure 10b shows the effect of applying Hanning-smoothing. Notice that single channel RFI has spread into three channels. This spreading of RFI to other channels will ultimately result in a little more data being flagged.
Automatic RFI Excision
Now that we're done with online flagging, we can move on to removing some of the RFI present with auto-flagging algorithms used within flagdata, TFCrop and rflag. For further details on the two algorithm based tasks we will employ, please see the RFI presentation given at the 5th VLA Data Reduction Workshop.
TFCrop
Task flagdata's TFCrop is an algorithm that detects outliers in the 2D time-frequency plane and can operate on uncalibrated (non-bandpass corrected) data. TFCrop will iterate through segments of time and undergo several steps in order to find and excise different types of RFI:
Step 1: Detect short-duration RFI spikes (narrow- and broad-band).
Step 2: Search for time-persistent RFI.
Step 3: Search for time-persistent, narrow-band RFI.
Step 4: Search for low-level wings of very strong RFI.
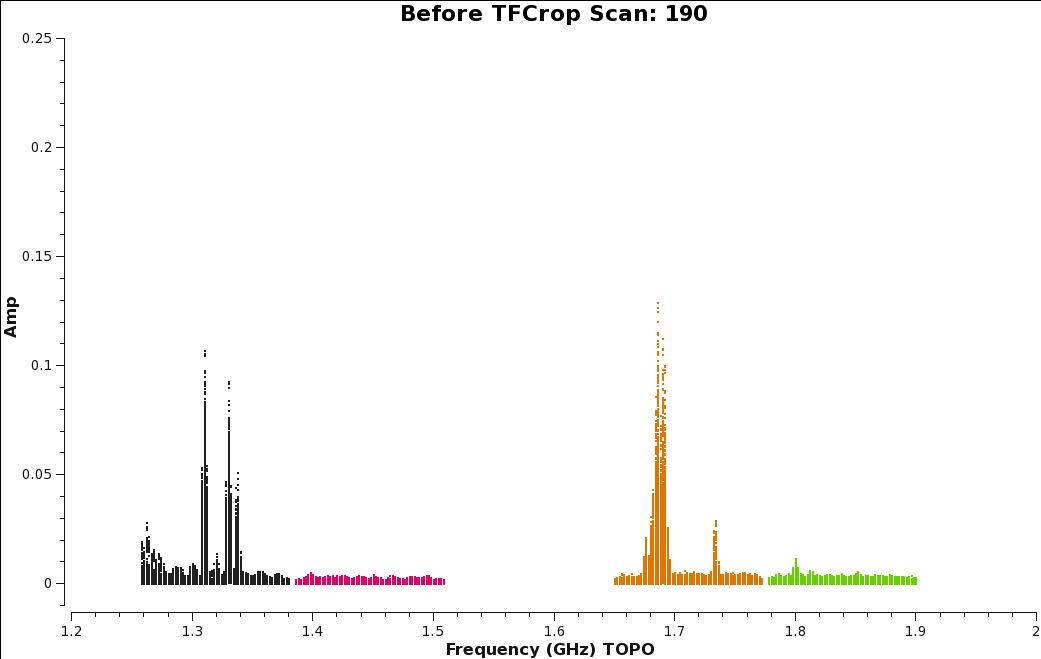
Amplitude vs. Frequency plot of Scan 190, before the TFCrop algorithm is applied.
We will apply the auto-flagging TFCrop algorithm for each spectral window. With TFCrop, it's a good idea to first inspect a small portion of data and review what and how much will be flagged. Once you are comfortable with the results, you can apply the algorithm to the remaining data. We will walk through the first spw and then include the remaining spws with the same command. First plot the corrected data for scan 190 (Figure 11) with all spectral windows so we can compare the data before and after TFCrop.
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='190', antenna='ea24', xaxis='freq', iteraxis='scan', yaxis='amp',
ydatacolumn='data', plotfile='amp_v_freq_before_tfcrop.png', title='Before TFCrop',
coloraxis='spw', plotrange=[1.2,2,-0.01,0.25])
The following are a set of flagdata commands which have been found to work reasonably well with these data. Take some time to play with the parameters, and the plotting capabilities, as this will be important when you are working with your own data later on. Because we set parameters display='both' and action='calculate', the flags are displayed but not actually written to the MS. This allows one to try different sets of parameters before actually applying the flags to the data.
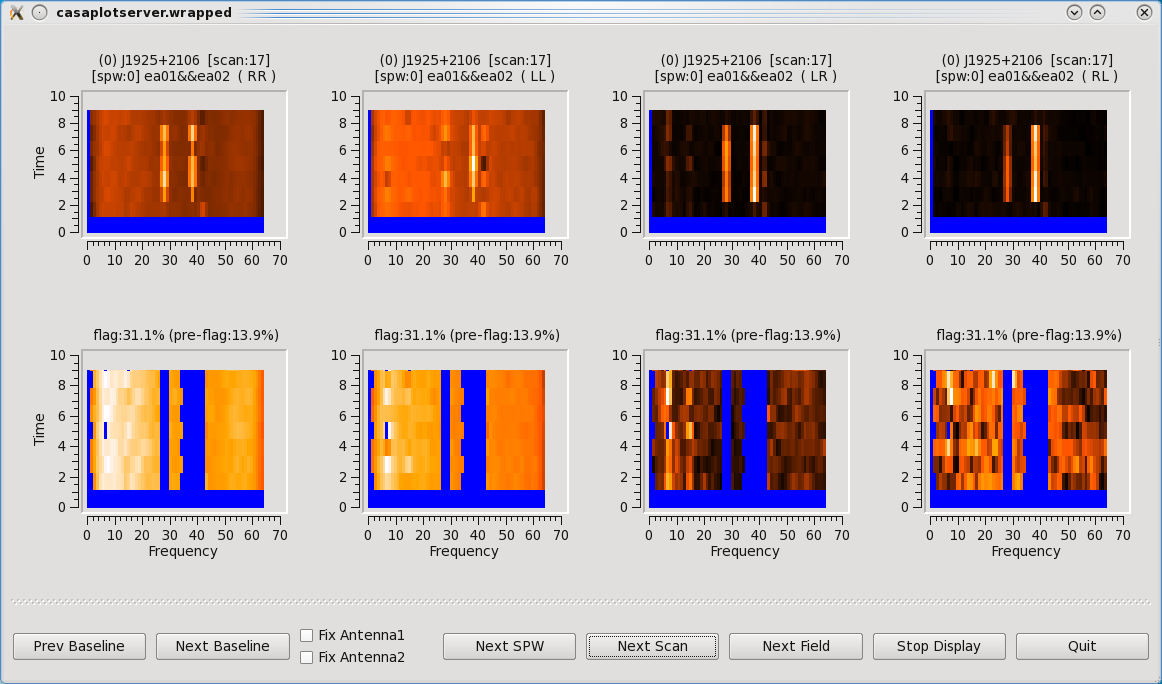
TFCrop flagging results. Top row is before, and bottom row is after TFCrop has applied flagging.
Some representative TFCrop plots are displayed (Figure 12). Each column displays an individual polarization product; all four polarizations are, from left to right, RR, RL, LR, and LL. The first row shows the data with current flags applied and the second includes the flags generated by flagdata. The x-axis is channel number (the spectral window ID is displayed in the top title) and the y-axis of the first two rows is all integrations included in a time segment, set by the parameter ntime. These are the data considered by the TFCrop algorithm during its flagging process, and changes in ntime will have some (relatively small) effect on what data are flagged.
Each plot page displays data for a single baseline and time segment, with colours representing the intensity of the visibilities (amplitude) in the heat colour scheme ranging from black for lowest amplitude points to white for highest amplitude data points. The flagged, or to be flagged, visibilities are masked in blue. The buttons at the bottom allow one to step through baseline (backward and forward), spw, scan, and field while Stop Display will continue the flagging operation without the GUI, and Quit aborts the run.
spw 0
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='tfcrop', spw='0',
datacolumn='data', action='calculate',
display='both', flagbackup=False)
The first two scans (14,15) are fully flagged (hence displayed as completely blue). Click on Next Scan until you reach scan 17 where it will be easier to see the flagging the algorithm performs. The top row shows current flags already applied to the data. The bottom row illustrates the flagging that can be applied with TFCrop, represented by the additional blue areas. Iterate through several scans and baselines. Once you're done, click on the Quit button on the bottom right corner. Let's now apply these flags by changing the action parameter.
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='tfcrop', spw='0',
datacolumn='data', action='apply',
display='', flagbackup=False)
The logger will report the percentage of flagged data in the table selection. We can now apply the TFCrop algorithm to the remaining spectral windows.
spw 1, 2, 3
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='tfcrop', spw='1~3',
datacolumn='data', action='apply',
display='both', flagbackup=False)
Iterate through several scans and baselines and, once you're ready to apply the flags, click on Stop Display. After TFCrop has gone through and flagged some of the worst RFI, we can inspect the log report and take note of how much has been flagged.
We can also use plotms to review the effects of using TFCrop.
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='190', antenna='ea24', xaxis='freq', iteraxis='scan', yaxis='amp',
ydatacolumn='data', plotfile='amp_v_freq_after_tfcrop.png', title='After TFCrop',
correlation='RR,LL', coloraxis='spw', plotrange=[1.2,2,-0.01,0.25])
Compare the plots before applying TFCrop (Figure 11) to the plot after applying TFCrop (Figure 13). Figure 13 shows great improvements, especially for spectral window 0 and 2, which had some of the worst RFI.
 Amplitude vs. Frequency plot of Scan 190, before the TFCrop algorithm is applied. |
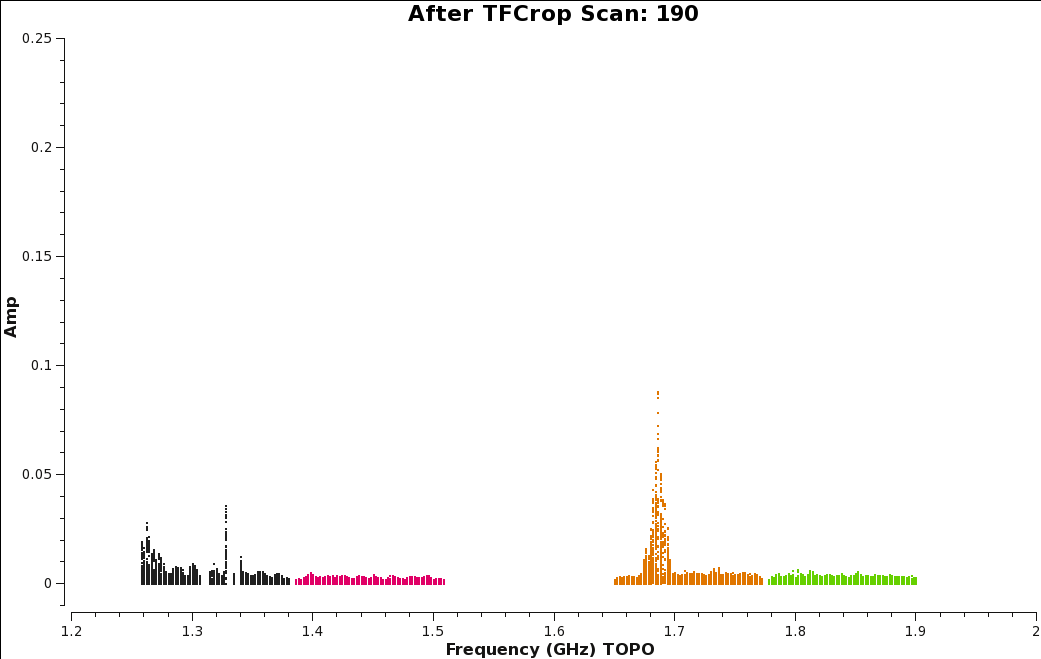 Plot of amplitude vs. frequency showing the effects of applying TFCrop to our data, for scan 190. |
Flagging Summary
We now calculate the amount of data flagged so far in our Measurement Set by using the flagdata task with parameter mode='summary' and assign the returned Python dictionary to the variable flagInfo.
# In CASA
flagInfo = flagdata(vis='SNR_G55_10s-hanning.ms', mode='summary')
Now we parse the dictionary to print some of the flagging statistics, first per source, then per spectral windows.
# In CASA
print("\n %2.1f%% of G55.7+3.4, %2.1f%% of 3C147, and %2.1f%% of J1925+2106 are flagged. \n" %
(100.0 * flagInfo['field']['G55.7+3.4']['flagged'] / flagInfo['field']['G55.7+3.4']['total'],
100.0 * flagInfo['field']['0542+498=3C147']['flagged'] / flagInfo['field']['0542+498=3C147']['total'],
100.0 * flagInfo['field']['J1925+2106']['flagged'] / flagInfo['field']['J1925+2106']['total']))
Note: IPython currently prevents copy/pasting several lines of code. To remedy this, we will use the %cpaste command to allow multiple lines to be placed in the terminal. Make sure you also copy the "--" characters to close the %cpaste input.
# In CASA
%cpaste
# Press Enter or Return, then copy/paste the following:
print("Spectral windows are flagged as follows:")
for spw in range(0,4):
print("SPW %s: %2.1f%%" % (spw, 100.0 * flagInfo['spw'][str(spw)]['flagged'] / flagInfo['spw'][str(spw)]['total']))
--
We can now move to the other auto-flagging algorithm, rflag.
RFlag
RFlag, like TFCrop, is an autoflag algorithm which uses a sliding window statistical filter. Data are iterated through in segments of time where statistics are accumulated and thresholds calculated.
WARNING: It is important to not assume that rflag can run on your target. This is especially important for spectral lines, as it may mistake them for interference.
IMPORTANT: Task rflag should be executed on calibrated data only. For more details, please see Emmanuel Momjian's presentation on VLA Data Reduction Techniques and Calibration given during the 5th VLA Data Reduction Workshop.
Bandpass calibration (to demonstrate RFlag)
For the purpose of demonstrating the functionality of RFlag on our data, we need to perform bandpass calibration first. Here, we just briefly outline the bandpass calibration process. For detailed calibration tutorial see VLA Data Reduction Tutorials.
First, run gaincal to solve for an initial set of antenna-based phases over a narrow range of channels. To find channels for each spectral window that are relatively RFI-free over the course of the observing session, look at the data with plotms. Use only a narrow channel range.
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='42,65,88,11,134,157', antenna='ea24', xaxis='channel',
yaxis='amp', iteraxis='spw')
Iterating over each spectral window plot, search for channel ranges with stable amplitudes. An example of a few appropriate channel ranges include: SPW 0:20~24, SPW 1:49~52, SPW 2:38~41, SPW 3:41~44.
Using these ranges, run gaincal to calculate phase-only solutions that will be used as input during the initial bandpass calibration. Remember: the calibration tables we are creating now are so that we can use an automatic RFI flagging algorithm. The final calibration tables will be generated later, after automated flagging. Here are the inputs for the initial pre-bandpass phase calibration:
# In CASA
gaincal(vis='SNR_G55_10s-hanning.ms', caltable='SNR_G55_10s-hanning.initPh', field='J1925+2106',
solint=' int ', spw='0:20~24,1:49~52,2:38~41,3:41~44', refant='ea24', minblperant=3,
minsnr=3.0, calmode='p')
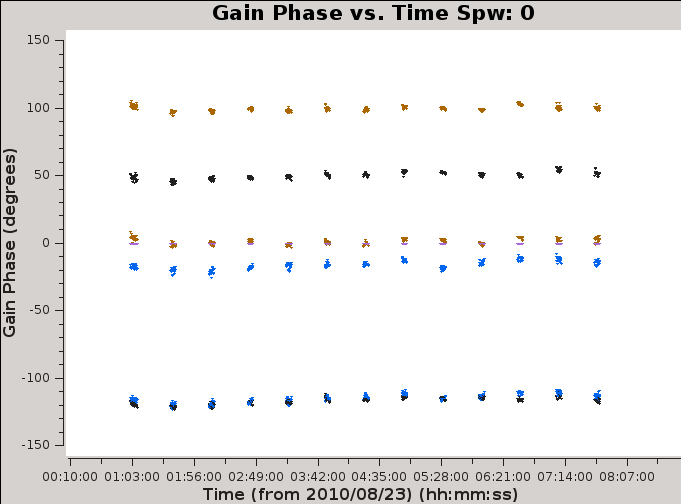
Gain Phase vs. Time for spw0, for four antennas.
It is always a good idea to inspect the resulting calibration table. We can do this with plotms.
# In CASA
plotms(vis='SNR_G55_10s-hanning.initPh', xaxis='time', yaxis='phase',
iteraxis='spw', antenna='ea01,ea05,ea10,ea24', coloraxis='Antenna1')
Figure 14 shows the phases do not change much over the course of the observing session.
Since the plots look fairly reasonable, we will now create a time-averaged bandpass solution for the phase calibration source using the bandpass task.
# In CASA
bandpass(vis='SNR_G55_10s-hanning.ms', caltable='SNR_G55_10s-hanning.initBP',
field='J1925+2106', solint=' inf ', combine='scan', refant='ea24',
minblperant=3, minsnr=10.0, gaintable='SNR_G55_10s-hanning.initPh',
interp='nearest', solnorm=False)
Now, inspect the resulting bandpass plots with plotms (Figures 15a,b,c).
# In CASA
plotms(vis='SNR_G55_10s-hanning.initBP',xaxis='freq', yaxis='amp', iteraxis='antenna',
coloraxis='corr',gridrows=2)
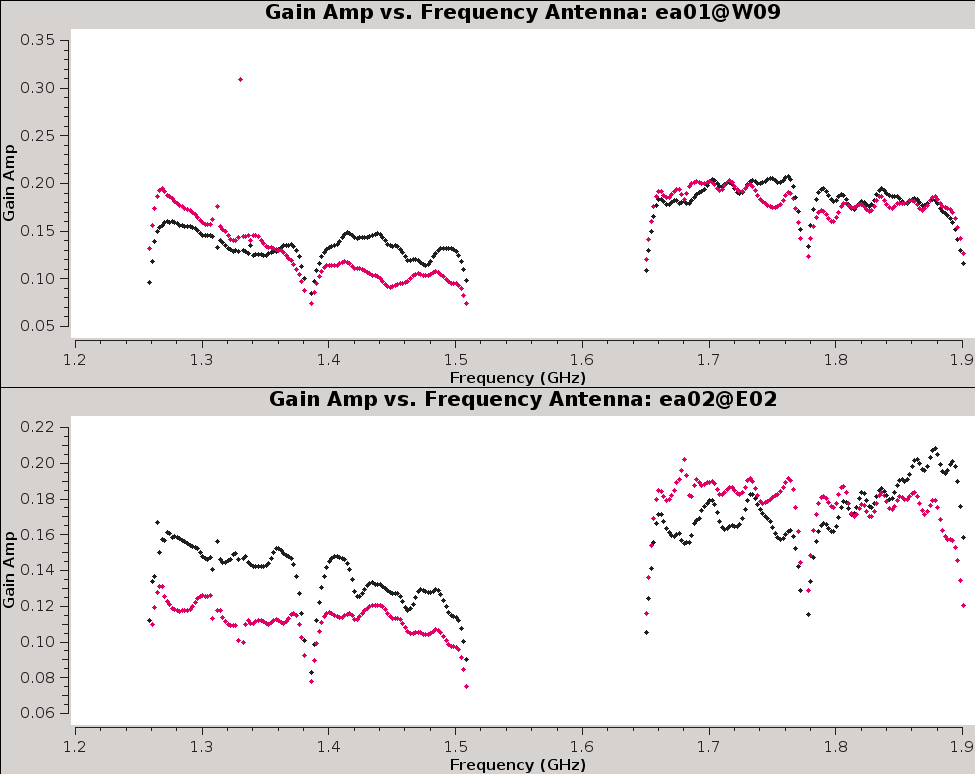 Bandpass for antennas ea01 and ea02. |
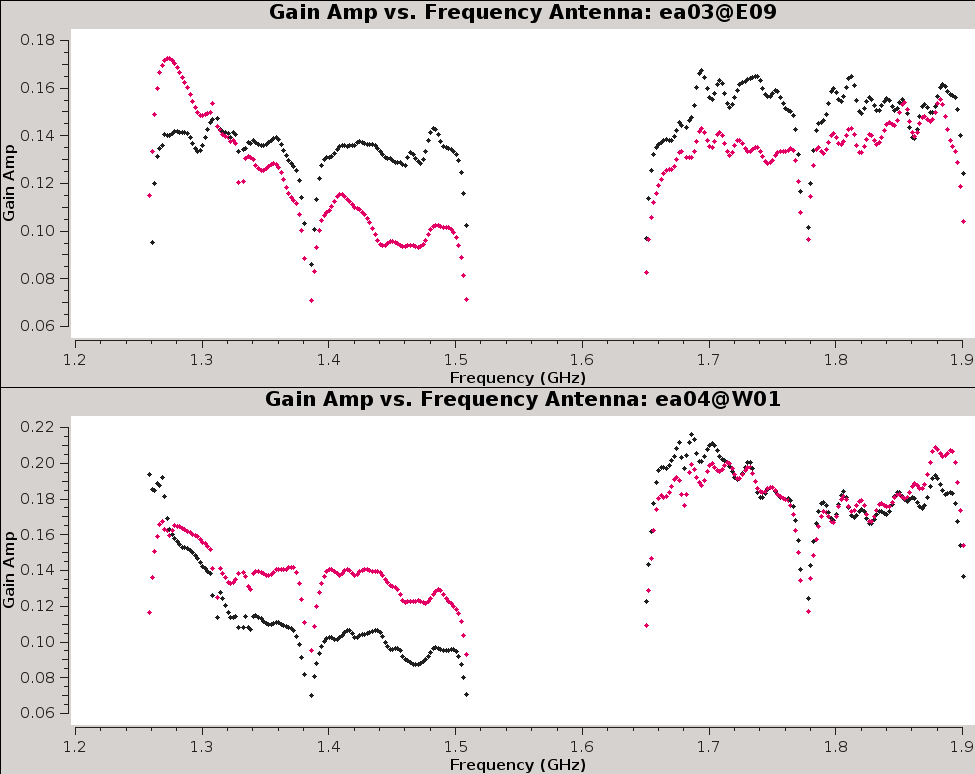 Bandpass for antennas ea03 and ea04. |
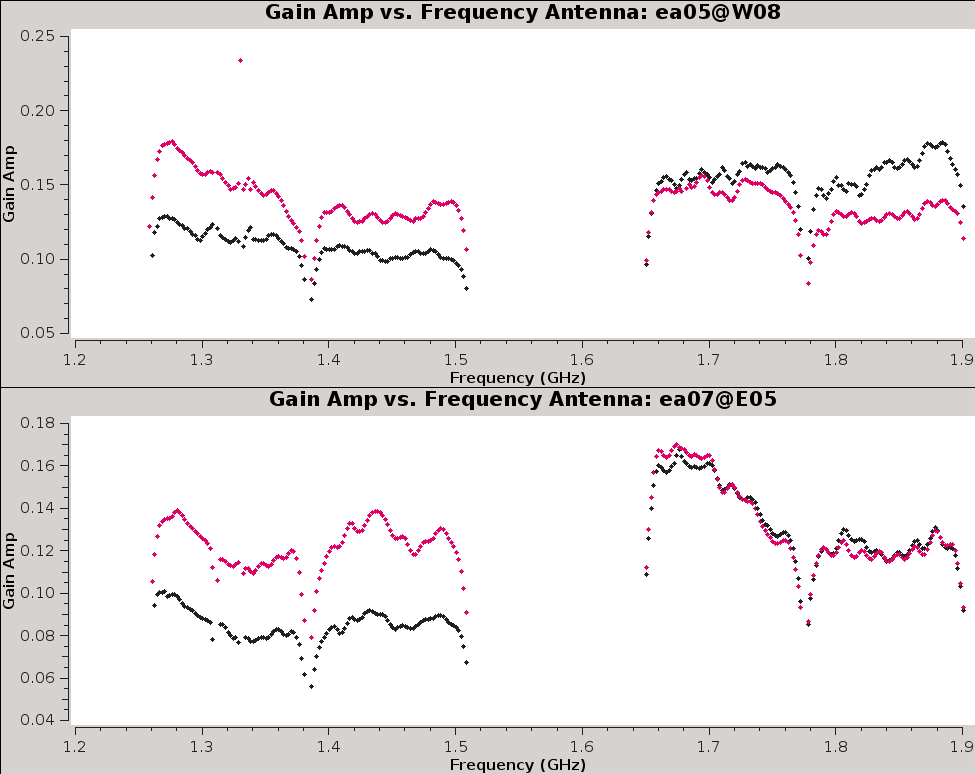 Bandpass for antennas ea05 - ea07 (note ea06 is flagged). |
Interactive flagging of bad solutions (calibration tables)
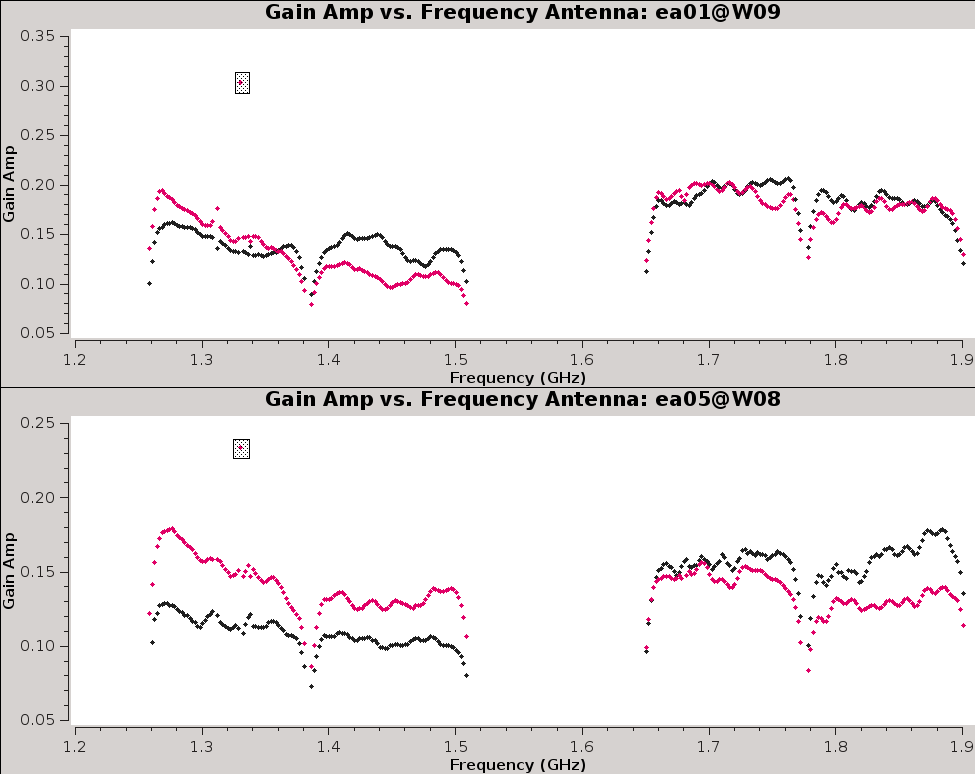
Interactive plotms flagging of the offset points for ea01 and ea05 in the calibration table.
We notice that antennas ea01 and ea05 have a point that is offset from the rest (Figures 15a,c). Let's plot just these two antennas, locate the point on the plot, and flag it interactively through plot. The interactive flagging within plotms will not create a backup, therefore it may be wise to use flagmanager before doing so (see Section Flagmanager earlier in this tutorial).
# In CASA
flagmanager(vis='SNR_G55_10s-hanning.initBP', mode='save', versionname='before_flagging')
# In CASA
plotms(vis='SNR_G55_10s-hanning.initBP',xaxis='freq', yaxis='amp', iteraxis='antenna',
antenna='ea01,ea05', coloraxis='corr', gridrows=2)
We will highlight the points by clicking on the Mark Region button, drawing boxes over the points, and clicking on the Flag button. Before doing this, one could also get information on the points by clicking on the Locate button, which will display details about the highlighted regions (Figure 16). After having flagged the offset points, your plots should update.
Applying calibration
We will now apply the bandpass calibration table using applycal.
# In CASA
applycal(vis='SNR_G55_10s-hanning.ms', gaintable='SNR_G55_10s-hanning.initBP', calwt=False)
This operation will flag data that correspond to flagged solutions, so applycal makes a backup version of the flags prior to operating on the data. Running this task might take a little while.
Running RFlag on bandpass calibrated data
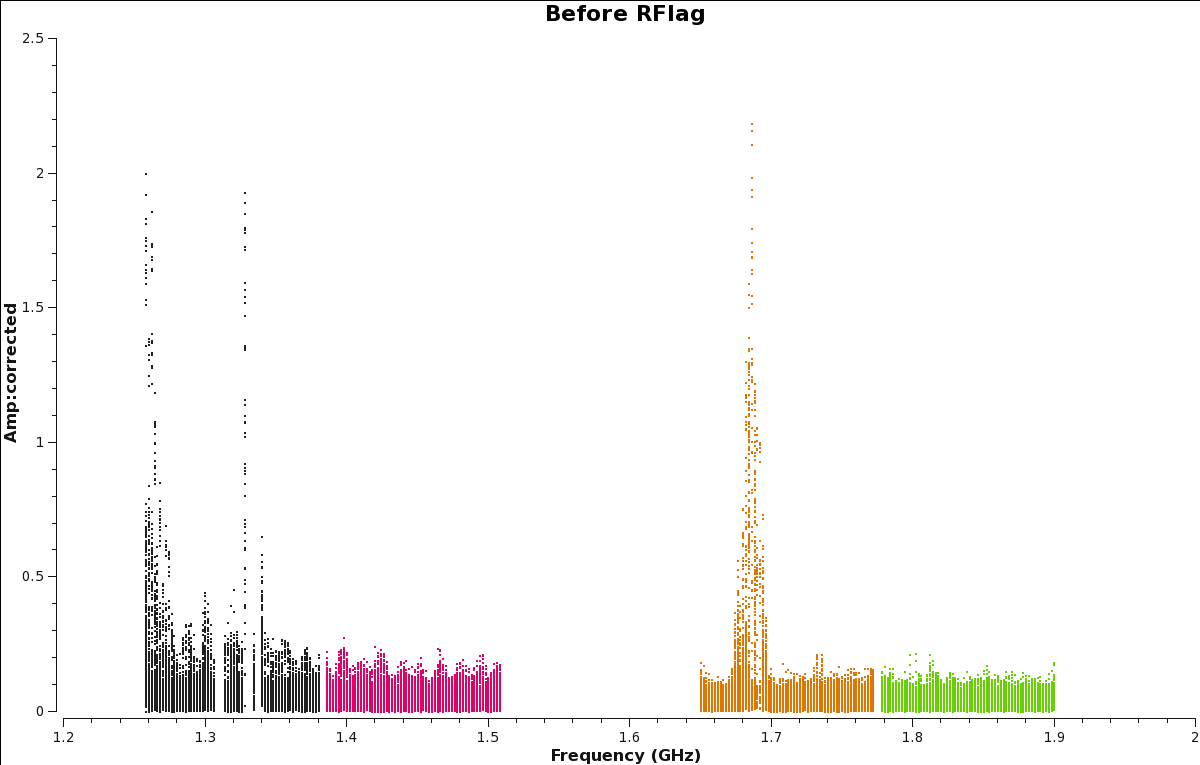
Amplitude vs. Frequency plot of scan 190, before RFlag is applied.
Now that we have bandpass-corrected data, we will run flagdata] in rflag mode.
We will use flagdata with parameters mode='rflag', and action='calculate' to first review the amount of data to be flagged. We will also change the parameter datacolumn='corrected' since we've applied the bandpass corrections to the MS and, in the process, created the corrected_data column in the MS table.
spw 0
First, let's create a plot of our corrected data before we apply RFlag (Figure 17).
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='190' , antenna='ea24', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', coloraxis='spw', title='Before RFlag',
plotfile='amp_v_freq.beforeRFlag.png')
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='rflag', spw='0', datacolumn='corrected',
action='calculate', display='both', flagbackup=False)
After reviewing the calculated flags, we can see that a lot of RFI is being missed (Figure 18). We will need to modify the default parameters for this spectral window. Click on Quit on the lower right hand side of the window. Let's review the parameters for flagdata and modify them:
# In CASA, get the last called parameter values for flagdata
tget flagdata
# In CASA, inspect the inputs
inp
The default timedevscale=5.0 and freqdevscale=5.0 parameters are not flagging enough bad data. We can provide more stringent values to the time/freq deviation scales parameters. A value smaller than the default (5.0) will flag more data in either the time or frequency axes, as can be seen on the plots in the displayed window. A value larger than the default will result in less flagged data.
Since we know spw 0 has lots of RFI, we will change parameters freqdevscale=2.5 and timedevscale=3.5. We will also change parameter action='apply'. Note that we can always decide that this still isn't good enough and click on Quit to change our parameter values (will not perform flagging). Once the window opens, proceed to scan 22 (Figure 19), review what will be flagged, and click on Stop Display (as we did during TFCrop) to allow the task to continue with the flagging.
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='rflag', spw='0', datacolumn='corrected',
freqdevscale=2.5, timedevscale=3.5, action='apply', display='both', flagbackup=False)
Although RFlag has done a pretty good job of finding the bad data, some still remains. One way to excise the remaining bad data is to use the parameter mode='extend' feature of flagdata, which can extend flags along a chosen axis and removes islands of small data patches in the midst of flagged data. We first extend the flags across polarization so if any one polarization is flagged, all data for that time / channel will be flagged:
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='extend', spw='0', extendpols=True,
action='apply', display='', flagbackup=False)
Now we extend the flags in time and frequency using the parameters growtime and growfreq. For this data, the rflag algorithm seems most likely to miss RFI that should be flagged along more of the time axis. We will try with parameter growtime=50.0, which will flag all data for a given channel if more than 50% of that channel's time is already flagged, and parameter growfreq=90.0, which will flag the entire spectrum for an integration if more than 90% of the channels in that integration are already flagged.
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', mode='extend', spw='0', growtime=50.0, growfreq=90.0,
action='apply', display='', flagbackup=False)
spw 1, 2, and 3
Now, let's work on SPW's 1,2, and 3. We've chosen parameter display='both' in order to first review the flags to decide if too much or too little is being flagged. We can always click on Quit and decide to change parameter values for freqdevscale and timedevscale as we did with spectral window 0. Note that spw 2 has more stringent parameter values compared to spw 1 and 3, as it contains more RFI. To allow the flagging to continue, click on Stop Display.
# In CASA, run RFlag on spectral windows 1 & 3
flagdata(vis='SNR_G55_10s-hanning.ms', mode='rflag', spw='1,3', datacolumn='corrected',
freqdevscale=4.0, timedevscale=4.0, action='apply', display='both', flagbackup=False)
# In CASA, extend flags in polarization for spectral windows 1 & 3
flagdata(vis='SNR_G55_10s-hanning.ms', mode='extend', spw='1,3', extendpols=True, action='apply',
display='', flagbackup=False)
# In CASA, extend flags in time and frequency for spectral windows 1 & 3
flagdata(vis='SNR_G55_10s-hanning.ms', mode='extend', spw='1,3', growtime=50.0, growfreq=90.0,
action='apply', display='', flagbackup=False)
# In CASA, run RFlag on spectral window 2 with more RFI, hence smaller frequency and time deviation scale values
flagdata(vis='SNR_G55_10s-hanning.ms', mode='rflag', spw='2', datacolumn='corrected',
freqdevscale=2.5, timedevscale=2.5, action='apply', display='both', flagbackup=False)
# In CASA, extend flags in polarization for spectral window 2
flagdata(vis='SNR_G55_10s-hanning.ms', mode='extend', spw='2', extendpols=True, action='apply',
display='', flagbackup=False)
# In CASA, extend flags in time and frequency for spectral window 2
flagdata(vis='SNR_G55_10s-hanning.ms', mode='extend', spw='2', growtime=50.0, growfreq=90.0,
action='apply', display='', flagbackup=False)
Let's create an after RFlag plot to see the improvements (Figure 20). Compare the resulting plot to the before TFCrop plot (Figure 17).
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='190' , antenna='ea24', xaxis='freq', yaxis='amp',
ydatacolumn='corrected', coloraxis='spw', title='After RFlag', plotfile='amp_v_freq.afterRFlag.png',
plotrange=[1.2,2,0,2.5])
 Amplitude vs. Frequency plot of scan 190, before RFlag is applied. |
 Plot of amplitude vs. frequency of scan 190, after having run RFlag. |
Interactive Flagging
To flag data interactively (manually), one can use plotms. We will be scanning for data points that look odd. Please note that flagging data through plotms will not create a backup, so it's important to use the flagmanager() before deciding to mark your regions for flagging purposes.
# In CASA
flagmanager(vis='SNR_G55_10s-hanning.ms', mode='save',
versionname='before_interactive_flagging')
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='50~100', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', coloraxis = 'spw')
Figure 21 (next section) shows some points in green that have higher amplitudes than the rest. Use the Mark Regions button (square with green plus sign) to highlight the region, and the Locate button (magnifying glass with white page) to obtain information on the data points. We can now decide to flag all of the highlighted area by clicking on the Flag button (red flag). The region is now flagged.
Similarly, one can also perform interactive flagging with msview. Again, the flagging is performed directly on the data, and so it is beneficial to execute flagmanager as above before embarking on interactive flagging with msview().
Open msview() and load the data as described in section Identifying RFI with MSview:
# In CASA
msview()
Select what you would like the axes to display (by default msview will plot time vs baseline), and using zoom tool (magnifying glass button with plus sign) zoom in on the data chunks you would like to inspect and potentially flag. Now, draw a region around the bad data using the drawing selectors (rectangle, circle, polygon) and right-double click to flag. To save the flagging, click Save Edits button in the flagging tab of Data Display Option window.
Flagging data interactively can be useful, but is discouraged, especially for large data volumes.
A more proper form of flagging would be to narrow your search on bad data points (Locate data as described above) and use a flagging task to remove them as showed in the next section below.
Identifying remaining bad data with plotms()/msview()
Once the bulk of the RFI and corrupted data has been removed with the methods described earlier in this guide, it is worth having another look at the data with the visualisation tools, either plotms() or msview(). This step may be valuable to perform once your data are calibrated and ready for imaging, to double check all the bad data has been indeed removed.
Let's again inspect amplitude vs uv-distance of our calibrated target data (Figure 21) with plotms(). Instead of flagging the data interactively (the Flag button), it is recommended to run the flagdata task. While locating the bad data points in the previous section (Mark Region and then click Locate button), CASA logger provided us with information on the data points. Notice that they all belong to scans 88 and 87. The scans also involve a baseline shared with ea16. To flag these data run the flagdata() task:
# In CASA
flagdata(vis='SNR_G55_10s-hanning.ms', scan='87,88', antenna='ea16', flagbackup=True)
To see the effect of this flagging, either Reload the image (tick Reload and click Plot) if your plotms() session from the previous section is still open, or otherwise run the following command:
# In CASA
plotms(vis='SNR_G55_10s-hanning.ms', scan='50~100', xaxis='uvdist', yaxis='amp',
ydatacolumn='corrected', coloraxis = 'spw')
We can now see the image (Figure 22) looks better without scans 87 and 88 for ea16.
If you would like to perform these steps with msview(), instead of plotms(), plot time vs. baseline length. Here baseline length is equivalent to the uv-distance.
Flagging Summary & Report
We will now run flagdata in summary mode to inspect how much total data has been flagged. We can also create a plot of the percentage of flagged data with respect to frequency by setting parameter display='report' (Figure 23). The report parameter will also create a plot of the antennas, with the circle size representing the percentage of flagged data per antenna (Figure 24). This can be viewed by clicking Next. One can also zoom in and out for a better view.
# In CASA
flagInfo = flagdata(vis='SNR_G55_10s-hanning.ms', mode='summary', action='calculate', display='report', spwchan=True)
# In CASA
print("\n %2.1f%% of G55.7+3.4, %2.1f%% of 3C147, and %2.1f%% of J1925+2106 are flagged. \n" %
(100.0 * flagInfo['field']['G55.7+3.4']['flagged'] / flagInfo['field']['G55.7+3.4']['total'],
100.0 * flagInfo['field']['0542+498=3C147']['flagged'] / flagInfo['field']['0542+498=3C147']['total'],
100.0 * flagInfo['field']['J1925+2106']['flagged'] / flagInfo['field']['J1925+2106']['total']))
# In CASA
%cpaste
# Press Enter or Return, then copy/paste the following:
print("Spectral windows are flagged as follows:")
for spw in range(0,4):
print("SPW %s: %2.1f%%" % (spw, 100.0 * flagInfo['spw'][str(spw)]['flagged'] / flagInfo['spw'][str(spw)]['total']))
--
As a result of the flagging we have removed over 40% of the data for G55.7+3.4 and, as expected, spectral windows 0 and 2 have the most flagged data.
In addition, we can inspect the CASA log, which will report the percentage of flagged data for:
1. per Spw/ per Channel
2. Each Antenna
3. Every Correlation (RR, RL, LL, LR)
4. Every Scan
5. Every spectral window
Last checked on CASA Version 5.4.0.