Jupiter: continuum polarization calibration 5.5.0: Difference between revisions
m Fschinze moved page Jupiter: continuum polarization calibration to Jupiter: continuum polarization calibration 5.5.0 |
|||
| (34 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
| style="background: red; color: white;" | '''DISCLAIMER''' | | style="background: red; color: white;" | '''DISCLAIMER''' | ||
|- | |- | ||
| This is an advanced reference guide to calibration and imaging of pre-upgrade VLA polarimetric data with CASA 5. | | This is an advanced reference guide to calibration and imaging of pre-upgrade VLA polarimetric data with CASA 5.5.0. If you are a beginning or novice user, please consult relevant [https://casaguides.nrao.edu/index.php/Karl_G._Jansky_VLA_Tutorials CASA guides] first. | ||
|} | |} | ||
| Line 13: | Line 10: | ||
== Data Import == | == Data Import == | ||
The data set for this tutorial lives [https://casa.nrao.edu/Data/VLA/Planets6cm/ here] (129MB). This is a VLA 6cm dataset that was observed back in 1999 to set the flux scale for calibration of the VLA. Included in the program were observations of the planets, including Jupiter which we will focus on in this tutorial. These are D-configuration data, with resolution of around 14". | The data set for this tutorial lives [https://casa.nrao.edu/Data/VLA/Planets6cm/planets_6cm.fits here: planets_6cm.fits] (129MB). This is a VLA 6cm dataset that was observed back in 1999 to set the flux scale for calibration of the VLA. Included in the program were observations of the planets, including Jupiter which we will focus on in this tutorial. These are D-configuration data, with resolution of around 14". | ||
Import the data into CASA. Task <tt>importuvfits()</tt> will read the original AIPS friendly UVFITS format and create a CASA native MS. | Import the data into CASA. Task <tt>importuvfits()</tt> will read the original AIPS friendly UVFITS format and create a CASA native MS. | ||
| Line 21: | Line 17: | ||
importuvfits(fitsfile='planets_6cm.fits', vis='jupiter6cm.demo.ms', antnamescheme='old') | importuvfits(fitsfile='planets_6cm.fits', vis='jupiter6cm.demo.ms', antnamescheme='old') | ||
</source> | </source> | ||
Check the observation set up and print verbose summary of the observations to the CASA logger | Check the observation set up and print verbose summary of the observations to the CASA logger | ||
| Line 28: | Line 23: | ||
listobs(vis='jupiter6cm.demo.ms') | listobs(vis='jupiter6cm.demo.ms') | ||
</source> | </source> | ||
The output of this task is fairly long, the abridged version is shown below since we may need some of this information later on during our calibration. | The output of this task is fairly long, the abridged version is shown below since we may need some of this information later on during our calibration. | ||
| Line 107: | Line 101: | ||
</source> | </source> | ||
<p style="color: #ff0000;"> <b>Note</b> (for uvfits sourced directly from archive only): CASA 5.4 and earlier versions will not plot the antennas properly. This is to be resolved in future CASA versions. If this occurs then a reference antenna can be selected using the listobs task and choosing an antenna located on a pad close to the center of the array (Pads numbers increase with distance from the center of the array, ie. "VLA:_N1" and "EVLA:W2" are pads close to the center of the array, where as, "VLA:_N18" is not).</p> | |||
<p style="color: #ff0000;"><b> | |||
== Data Inspection and Editing == | == Data Inspection and Editing == | ||
| Line 137: | Line 128: | ||
<source lang="python"> | <source lang="python"> | ||
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='1331+305',correlation=' | flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='1331+305',correlation='ABS_RR,LL',clipoutside=False,clipminmax=[0,0.75]) | ||
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='1331+305',correlation=' | flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='1331+305',correlation='ABS_RL,LR',clipoutside=False,clipminmax=[0,0.04]) | ||
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='0137+331',correlation=' | flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='0137+331',correlation='ABS_RR,LL',clipoutside=False,clipminmax=[0,0.55]) | ||
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='0137+331',correlation=' | flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='0137+331',correlation='ABS_RL,LR',clipoutside=False,clipminmax=[0,0.01]) | ||
</source> | </source> | ||
If you are unfamiliar with flagging in CASA, consult detailed topical guide [https://casaguides.nrao.edu/index.php/VLA_CASA_Flagging-CASA5.0.0 Flagging VLA Data]. | If you are unfamiliar with flagging in CASA, consult detailed topical guide [https://casaguides.nrao.edu/index.php/VLA_CASA_Flagging-CASA5.0.0 Flagging VLA Data]. | ||
| Line 148: | Line 139: | ||
== Calibration == | == Calibration == | ||
=== | === Flux Density Scale === | ||
Set the absolute flux density scale, but only for Stokes I at the moment (total flux density model). Our primary flux calibrator here is 1331+305 (3C286). The default model for CASA 5. | Set the absolute flux density scale, but only for Stokes I at the moment (total flux density model). Our primary flux calibrator here is 1331+305 (3C286). The default model for CASA 5.5+ is 'Perley-Butler 2017'. | ||
<source lang="python"> | <source lang="python"> | ||
| Line 175: | Line 166: | ||
# And check the solutions | # And check the solutions | ||
plotms(vis='jupiter6cm.demo.G', xaxis='time', yaxis='amp', gridrows=3, gridcols=3, iteraxis='antenna') | |||
plotms(vis='jupiter6cm.demo.G', xaxis='time', yaxis='phase', plotrange=[-1,-1,-200,200], gridrows=3, gridcols=3, iteraxis='antenna') | |||
</source> | </source> | ||
| Line 201: | Line 192: | ||
<source lang="python"> | <source lang="python"> | ||
plotms(vis='jupiter6cm.demo.Gflx', xaxis='time', yaxis='amp', showgui=True, plotfile='jupiter6cm.demo.Gflx.amp.png') | |||
plotms(vis='jupiter6cm.demo.Gflx', xaxis='time', yaxis='phase', plotrange=[-1,-1,-200,200], showgui=True, plotfile='jupiter6cm.demo.Gflx.phase.png') | |||
</source> | </source> | ||
| Line 238: | Line 229: | ||
'format': "{field Id: {spw Id: {fluxd: [I,Q,U,V] in Jy}, 'fieldName':field name }}"} | 'format': "{field Id: {spw Id: {fluxd: [I,Q,U,V] in Jy}, 'fieldName':field name }}"} | ||
</pre> | </pre> | ||
==== Solve for the leakage terms (D terms) ==== | ==== Solve for the leakage terms (D terms) ==== | ||
| Line 250: | Line 242: | ||
<source lang="python"> | <source lang="python"> | ||
plotms(vis='jupiter6cm.demo.D', xaxis='antenna', yaxis='amp', showgui=True, plotfile='jupiter6cm.demo.D.amp.png') | |||
plotms(vis='jupiter6cm.demo.D', xaxis='antenna', yaxis='phase', plotrange=[-1,-1,-200,200], showgui=True, plotfile='jupiter6cm.demo.D.phase.png') | |||
plotms(vis='jupiter6cm.demo.D', xaxis='antenna', yaxis='snr', showgui=True, plotfile='jupiter6cm.demo.D.snr.png') | |||
</source> | </source> | ||
| Line 279: | Line 272: | ||
<!--<p style="color: #ff0000;"><b> CASA 5.3 bug (present from CASA 5.0+; reported as JIRA CAS-11762). </b> For the time being please use CASA <=4.7, split the data set into single SPW (IF) MS files, and calibrate them separately, or just calibrate each spw separately by specifying, in each task, spw='x' where x is whichever spw you want.</p>--> | <!--<p style="color: #ff0000;"><b> CASA 5.3 bug (present from CASA 5.0+; reported as JIRA CAS-11762). </b> For the time being please use CASA <=4.7, split the data set into single SPW (IF) MS files, and calibrate them separately, or just calibrate each spw separately by specifying, in each task, spw='x' where x is whichever spw you want.</p>--> | ||
<b>NOTE</b>: If you are using CASA 4.7 or earlier, you may want to use <tt>poltype='X'</tt> in X-term calculations (Mueller matrices). For CASA 5.0 and later this option will not work (the relevant CASA documentation is being updated), and hence you need to use <tt>poltype='Xf'</tt> (Jones matrices). Although 'Xf' should be used predominantly for large bandwidths, which clearly is not the case here, it can also be used for the single channel old VLA data as long as you make sure the <tt>interp</tt> parameter is set to default (which should be <tt>nearest</tt>). | |||
=== Apply the calibration === | === Apply the calibration === | ||
| Line 301: | Line 296: | ||
You can use <tt>plotms()</tt> task to look at the split calibrated data. | You can use <tt>plotms()</tt> task to look at the split calibrated data. | ||
== Initial imaging (Stokes I) == | == Initial imaging (Stokes I) == | ||
We will now image and clean the calibrated Jupiter data. In the next step we will perform self-calibration, so here in the initial imaging we will not clean too deeply and we will also save the model in the MS data (with the use of parameter <tt> | We will now image and clean the calibrated Jupiter data. In the next step we will perform self-calibration, so here in the initial imaging we will not clean too deeply and we will also save the model in the MS data (with the use of parameter <tt>savemodel</tt> in <tt>tclean()</tt>). | ||
<!-- | |||
<b>NOTE</b>: As of CASA 5.3 the recommended task for deconvolution is <tt>tclean()</tt>. However, we found there may be a problem with how <tt>tclean()</tt> handles old, pre-upgrade VLA data. This is currently being investigated, and once resolved, this guide will be updated to demonstrate the <tt>tclean()</tt> use in this data reduction. Meanwhile, please use the standard <tt>clean()</tt> for deconvolution and imaging. | |||
Check JIRA Ticket CAS-11971 for details regarding this comment. | |||
--> | |||
Since we set mask="" in our cleaning tasks, the interactive GUI will not allow you to do the deconvolution before you draw the mask on at least one plane. When drawing a mask, make sure to extend it to all channels and all polarisations. | |||
<source lang="python"> | <source lang="python"> | ||
tclean(vis='jupiter6cm.demo.JUPITER.split.ms', stokes='I', field='', spw='', imagename='jupiter6cm.demo.JUPITER.I.clean1', robust = 0.0, imsize=[288], cell=['3arcsec'], specmode='mfs', deconvolver='hogbom', weighting='briggs', threshold='0.1mJy', mask='', niter=500, interactive=True, cycleniter=100, savemodel='modelcolumn', pblimit=-0.2) | |||
</source> | </source> | ||
Inspect the quality of the cleaned images with <tt>imstat()</tt> | Inspect the quality of the cleaned images with <tt>imstat()</tt>. | ||
imstat(imagename='jupiter6cm.demo.JUPITER.I. | <source lang="python"> | ||
imstat(imagename='jupiter6cm.demo.JUPITER.I.clean1.image', stokes='I', box='216,1,287,72') | |||
</source> | |||
In this initial imaging the deconvolution will stop with the iteration limit since we set it fairly low. We 'cleaned' approximately 4.3 Jy in this initial imaging step, and reached rms of about 3.52 mJy/bm. | |||
<!-- Either of the tasks 'cleaned' approximately 4.3 Jy in this initial imaging step, and we get rms of about 3.52 mJy/bm (using <tt>clean()</tt>).<!-- and 3.9 mJy/bm (using <tt>tclean()</tt>).--> | |||
== Self-calibration == | == Self-calibration == | ||
| Line 335: | Line 332: | ||
In the self calibration step we run <tt>gaincal()</tt> that will make use of the newly created MODEL column in our MS file. | In the self calibration step we run <tt>gaincal()</tt> that will make use of the newly created MODEL column in our MS file. | ||
<source lang="python"> | |||
gaincal(vis='jupiter6cm.demo.JUPITER.split.ms', caltable='jupiter6cm.demo.JUPITER.split.selfcal1', field='', spw='', gaintype='G', calmode='p', solint='30s', combine='', refant='11', parang=False, append=False, selectdata=False) | gaincal(vis='jupiter6cm.demo.JUPITER.split.ms', caltable='jupiter6cm.demo.JUPITER.split.selfcal1', field='', spw='', gaintype='G', calmode='p', solint='30s', combine='', refant='11', parang=False, append=False, selectdata=False) | ||
</source> | |||
Inspect the solutions | Inspect the solutions | ||
<source lang="python"> | |||
plotms(vis='jupiter6cm.demo.JUPITER.split.selfcal1',xaxis='time',yaxis='amp',gridrows=2, gridcols=3, iteraxis='antenna') | |||
plotms(vis='jupiter6cm.demo.JUPITER.split.selfcal1',xaxis='time',yaxis='phase', gridrows=2, gridcols=3 ,iteraxis='antenna',plotrange=[-1,-1,-180,180]) | |||
</source> | |||
And apply the calibration to the DATA(creating a CORRECTED column in the MS) | |||
<source lang="python"> | |||
applycal(vis='jupiter6cm.demo.JUPITER.split.ms', field='', spw='', selectdata=False, gaintable=['jupiter6cm.demo.JUPITER.split.selfcal1'], gainfield=[''], interp=['nearest'],calwt=[False], applymode='calflag') | |||
</source> | |||
Now, image the self-calibrated data | |||
<source lang="python"> | |||
tclean(vis='jupiter6cm.demo.JUPITER.split.ms', stokes='I', field='', spw='', imagename='jupiter6cm.demo.JUPITER.I.clean2', robust= 0.0, imsize=[288], cell=['3arcsec'], specmode='mfs', deconvolver='hogbom', weighting='briggs', threshold='0.05mJy', mask='', niter=10000, interactive=True, cycleniter=100, pblimit=-0.2) | |||
</source> | |||
Check the image statistics | |||
<source lang="python"> | <source lang="python"> | ||
imstat(imagename='jupiter6cm.demo.JUPITER.I.clean2.image', stokes='I', box='216,1,287,72') | |||
</source> | </source> | ||
After one cycle of self-calibration with <tt>tclean()</tt> we reached rms=1.10 mJy/bm. But bear in mind that the rms values may differ for you since they will depend on how deeply you cleaned and how good a mask you applied. | |||
== Full Stokes Imaging == | |||
At this stage we will perform full Stokes imaging to create image of each Stokes parameter that we will later use to create polarised intensity and angle images. | |||
<source lang="python"> | <source lang="python"> | ||
tclean(vis='jupiter6cm.demo.JUPITER.split.ms', stokes='IQUV', field='', spw='', imagename='jupiter6cm.demo.JUPITER.IQUV.clean', robust= 0.0, imsize=[288], cell=['3arcsec'], specmode='mfs', deconvolver='clarkstokes', weighting='briggs', threshold='0.05mJy', mask='', niter=10000, interactive=True, cycleniter=100, pblimit=-0.2) | |||
</source> | </source> | ||
Inspect the quality of the cleaned images with <tt>imstat()</tt>, | Inspect the quality of the cleaned images with <tt>imstat()</tt> (do it for each Stokes separately, as otherwise imstat() will return an average of all Stokes). You should be reaching values of about 1.1 mJy/bm (I), 0.62 mJy/bm (Q), 0.90 mJy/bm (U), 0.16 mJy/bm (V). | ||
== Polarisation Intensity and Angle Images == | == Polarisation Intensity and Position Angle Images == | ||
[[Image:Jupiter-poli.png|thumb|Polarised intensity (POLI) image of Jupiter with overlaid polarised angle vectors (POLA). ]] | |||
[[Image:Jupiter-oldVLA-stokesI.png|thumb|Total continuum intensity (Stokes I) image of Jupiter with overlaid contours of its polarised intensity (POLI).]] | |||
We now want to save each Stokes plane as separate images. We can use either the <tt>imsubimage()</tt> or <tt>immath()</tt> tasks to do so. | We now want to save each Stokes plane as separate images. We can use either the <tt>imsubimage()</tt> or <tt>immath()</tt> tasks to do so. | ||
| Line 392: | Line 405: | ||
Finally, we want to create the polarization intensity and position angle | Finally, we want to create the polarization intensity (POLI) and position angle (POLA) images. | ||
<source lang="python"> | <source lang="python"> | ||
immath(imagename=['JUPITER.Q.image','JUPITER.U.image'], mode='poli', outfile='JUPITER.POLI.image') | immath(imagename=['JUPITER.Q.image','JUPITER.U.image'], mode='poli', outfile='JUPITER.POLI.image') | ||
immath(imagename=['JUPITER.Q.image','JUPITER.U.image'], mode='pola', outfile='JUPITER.POLA.image',polithresh=' | </source> | ||
We will now use the <tt>polithresh</tt> parameter when creating the polarisation angle image to get rid of the noise. First check the statistic in the POLI image, and then use it as a threshold while creating the POLA image (in the example below equal to 4 sigma). | |||
<source lang="python"> | |||
imstat(imagename='JUPITER.POLI.image', box='216,1,287,72') | |||
</source> | |||
<source lang="python"> | |||
immath(imagename=['JUPITER.Q.image','JUPITER.U.image'], mode='pola', outfile='JUPITER.POLA.image',polithresh='4.5mJy/beam') | |||
</source> | </source> | ||
| Line 403: | Line 426: | ||
And that's it. For the tutorial on how to perform analysis, display and manipulation of the polarisation images now, consult the appropriate [https://casaguides.nrao.edu/index.php/VLA_Continuum_Tutorial_3C391-CASA5.0.0 VLA CASA guides]. | |||
{{Checked 5.5.0-149}} | |||
[[Pre-upgrade VLA Tutorials | ↵ '''Pre-upgrade VLA Tutorials''']] | [[Pre-upgrade VLA Tutorials | ↵ '''Pre-upgrade VLA Tutorials''']] | ||
<!--Last edited by Trent Seelig, 2019-08-27--> | |||
Latest revision as of 17:00, 13 February 2023
| DISCLAIMER |
| This is an advanced reference guide to calibration and imaging of pre-upgrade VLA polarimetric data with CASA 5.5.0. If you are a beginning or novice user, please consult relevant CASA guides first. |
Data Import
The data set for this tutorial lives here: planets_6cm.fits (129MB). This is a VLA 6cm dataset that was observed back in 1999 to set the flux scale for calibration of the VLA. Included in the program were observations of the planets, including Jupiter which we will focus on in this tutorial. These are D-configuration data, with resolution of around 14".
Import the data into CASA. Task importuvfits() will read the original AIPS friendly UVFITS format and create a CASA native MS.
importuvfits(fitsfile='planets_6cm.fits', vis='jupiter6cm.demo.ms', antnamescheme='old')
Check the observation set up and print verbose summary of the observations to the CASA logger
listobs(vis='jupiter6cm.demo.ms')
The output of this task is fairly long, the abridged version is shown below since we may need some of this information later on during our calibration.
########################################## ##### Begin Task: listobs ##### Observer: FLUX99 Project: Observation: VLA Computing scan and subscan properties... Data records: 2021424 Total elapsed time = 85136.5 seconds Observed from 15-Apr-1999/23:15:25.0 to 16-Apr-1999/22:54:21.6 (TAI) [...] Fields: 13 ID Code Name RA Decl Epoch SrcId nRows 0 A 0137+331 01:37:41.299500 +33.09.35.13400 J2000 0 72946 1 T 0813+482 08:13:36.051800 +48.13.02.26200 J2000 1 227524 2 A 0542+498 05:42:36.137900 +49.51.07.23400 J2000 2 227058 3 T 0437+296 04:37:04.174700 +29.40.15.13600 J2000 3 113424 4 VENUS 04:06:54.109428 +22.30.35.90609 J2000 4 121098 5 A 0521+166 05:21:09.886000 +16.38.22.05200 J2000 5 81258 6 T 1411+522 14:11:20.647700 +52.12.09.14100 J2000 6 243608 7 A 1331+305 13:31:08.288100 +30.30.32.96000 J2000 7 307958 8 MARS 14:21:41.365747 -12.21.49.45444 J2000 8 118570 9 NGC7027 21:07:01.593000 +42.14.10.18600 J2000 9 136082 10 NEPTUNE 20:26:01.136316 -18.54.54.21127 J2000 10 157736 11 URANUS 21:15:42.828572 -16.35.05.59272 J2000 11 99412 12 JUPITER 00:55:34.043951 +04.45.44.70633 J2000 12 114750 Spectral Windows: (2 unique spectral windows and 1 unique polarization setups) SpwID Name #Chans Frame Ch0(MHz) ChanWid(kHz) TotBW(kHz) CtrFreq(MHz) Corrs 0 none 1 TOPO 4885.100 50000.000 50000.0 4885.1000 RR RL LR LL 1 none 1 TOPO 4835.100 50000.000 50000.0 4835.1000 RR RL LR LL Sources: 26 ID Name SpwId RestFreq(MHz) SysVel(km/s) 0 0137+331 0 0 0 0 0137+331 1 0 0 1 0813+482 0 0 0 1 0813+482 1 0 0 2 0542+498 0 0 0 2 0542+498 1 0 0 3 0437+296 0 0 0 3 0437+296 1 0 0 4 VENUS 0 0 0 4 VENUS 1 0 0 5 0521+166 0 0 0 5 0521+166 1 0 0 6 1411+522 0 0 0 6 1411+522 1 0 0 7 1331+305 0 0 0 7 1331+305 1 0 0 8 MARS 0 0 0 8 MARS 1 0 0 9 NGC7027 0 0 0 9 NGC7027 1 0 0 10 NEPTUNE 0 0 0 10 NEPTUNE 1 0 0 11 URANUS 0 0 0 11 URANUS 1 0 0 12 JUPITER 0 0 0 12 JUPITER 1 0 0 Antennas 27 [....] ##### End Task: listobs ##### ##########################################
Check also the array configuration and pick a reference antenna. Here, we will use antenna '11' as a reference since it's located in the middle of the array.
plotants(vis='jupiter6cm.demo.ms', figfile='jupiter6cm.demo.ant.png')
Note (for uvfits sourced directly from archive only): CASA 5.4 and earlier versions will not plot the antennas properly. This is to be resolved in future CASA versions. If this occurs then a reference antenna can be selected using the listobs task and choosing an antenna located on a pad close to the center of the array (Pads numbers increase with distance from the center of the array, ie. "VLA:_N1" and "EVLA:W2" are pads close to the center of the array, where as, "VLA:_N18" is not).
Data Inspection and Editing
To inspect and interactively flag bad data use task plotms(). Note: it is not recommended to use plotms() to interactively flag the new post-upgrade VLA data due to their large volume and large bandwidths. However, the old VLA data are much smaller and plotms() is capable of managing it interactively.
First, inspect the primary flux and polarisation angle calibrator, 1331+305 (3C286). Inspect RR,LL and RL,LR correlations separately due to large amplitude differences between them (when all correlations are plotted together, the RL,LR will seem like bad data due to their much lower amplitudes as compared to RR,LL). Use Mark Regions within the plotms() GUI to draw boxes around points to flag, and hit Flag to flag.
plotms(vis='jupiter6cm.demo.ms', selectdata=True, field='1331+305', correlation='RR,LL', xaxis='uvdist', yaxis='amp')
Now, inspect the other two sources we are interested in, the polarisation leakage calibrator (0137+331) and our target source (JUPITER).
Flagging can also be done non-interactively as per currently recommended CASA practise. Use task flagdata() to do so. For example, from inspecting our data in plotms() you should have noticed that Antenna 9 (ID=8) in spw 1 is often bad. The bad data on Antenna 9 are in the last 4 scans in spw='1' for the 0137+331 calibrator and for our target source (JUPITER), as well as in the last scan for all antennas in both spws on the target source. We can flag these points with the following flagdata() task executions.
flagdata(vis='jupiter6cm.demo.ms', mode='manual', field='0137+331', spw='1', antenna='9', timerange='42:00:00~48:00:00')
flagdata(vis='jupiter6cm.demo.ms', mode='manual', field='JUPITER', spw='1', antenna='9', timerange='16:26:00~22:20:00')
flagdata(vis='jupiter6cm.demo.ms', mode='manual', field='JUPITER', spw='', timerange='21:40:00~22:20:00')
There are more data points to flag -- keep flagging until you are happy with the results. The following commands will get rid of most bad data, but depending on how clean the data you want to proceed with, you may still want to inspect them in plotms() and flag interactively the reminder of bad datapoints.
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='1331+305',correlation='ABS_RR,LL',clipoutside=False,clipminmax=[0,0.75])
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='1331+305',correlation='ABS_RL,LR',clipoutside=False,clipminmax=[0,0.04])
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='0137+331',correlation='ABS_RR,LL',clipoutside=False,clipminmax=[0,0.55])
flagdata(vis='jupiter6cm.demo.ms',mode='clip',field='0137+331',correlation='ABS_RL,LR',clipoutside=False,clipminmax=[0,0.01])
If you are unfamiliar with flagging in CASA, consult detailed topical guide Flagging VLA Data.
Calibration
Flux Density Scale
Set the absolute flux density scale, but only for Stokes I at the moment (total flux density model). Our primary flux calibrator here is 1331+305 (3C286). The default model for CASA 5.5+ is 'Perley-Butler 2017'.
setjy(vis='jupiter6cm.demo.ms', field='1331+305', model='3C286_C.im')
Initial gain calibration
At this stage the data have an overall flux density scaling determined, but full gain solutions aren't there yet. The relevant task is gaincal() (analogous to the AIPS task CALIB). Gaincal() will produce a separate table with solutions, and we will use appropriate extensions to keep track of what is what.
NOTE: Since we have only two single-channel continuum spw, we do not want to do bandpass calibration nor solve for delays within spws as is currently done with wide bandwidth post-upgrade VLA data.
Firstly, generate an antenna zenith-angle dependent VLA gain curve calibration table
gencal(vis='jupiter6cm.demo.ms', caltable='jupiter6cm.demo.gc', caltype='gc')
Now, solve for antenna gains on 1331+305 and 0137+331, using the generated gain curve table (.gc).
gaincal(vis='jupiter6cm.demo.ms', caltable='jupiter6cm.demo.G', field='1331+305,0137+331', spw='', gaintype='G', calmode='ap', solint='inf', combine='', refant='11', minsnr=3, gaintable=['jupiter6cm.demo.gc'], parang=False)
# And check the solutions
plotms(vis='jupiter6cm.demo.G', xaxis='time', yaxis='amp', gridrows=3, gridcols=3, iteraxis='antenna')
plotms(vis='jupiter6cm.demo.G', xaxis='time', yaxis='phase', plotrange=[-1,-1,-200,200], gridrows=3, gridcols=3, iteraxis='antenna')
If all looks good, bootstrap the flux density scale of the flux calibrator onto the phase calibrators (CASA's fluxscale() task is equivalent to GETJY in AIPS). When executing fluxscale(), the calibration table with the extension .G is modified and stored as a new table with the extension .Gflx.
myFluxscale = fluxscale(vis='jupiter6cm.demo.ms', caltable='jupiter6cm.demo.G', fluxtable='jupiter6cm.demo.Gflx', reference='1331+305', transfer='0137+331', incremental=True, append=False, display=False)
The output is displayed in the logger as well as stored in the myFluxscale python dictionary.
Beginning fluxscale--(MSSelection version)------- Found reference field(s): 1331+305 Found transfer field(s): 0137+331 Flux density for 0137+331 in SpW=0 (freq=4.8851e+09 Hz) is: 5.29665 +/- 0.00449217 (SNR = 1179.09, N = 54) Flux density for 0137+331 in SpW=1 (freq=4.8351e+09 Hz) is: 5.34890 +/- 0.00176819 (SNR = 3025.07, N = 54) Fitted spectrum for 0137+331 with fitorder=1: Flux density = 5.32271 +/- 1.70229e-07 (freq=4.86004 GHz) spidx=-0.954124 (degenerate) Storing result in jupiter6cm.demo.Gflx Writing solutions to table: jupiter6cm.demo.Gflx
Before proceeding, inspect the flux density calibration and save results to a file.
plotms(vis='jupiter6cm.demo.Gflx', xaxis='time', yaxis='amp', showgui=True, plotfile='jupiter6cm.demo.Gflx.amp.png')
plotms(vis='jupiter6cm.demo.Gflx', xaxis='time', yaxis='phase', plotrange=[-1,-1,-200,200], showgui=True, plotfile='jupiter6cm.demo.Gflx.phase.png')
Polarisation calibration
Just as in the step of the initial gain calibration, since our old VLA data have single-channel continuum spws, we do not want to solve for KCROSS delays in our polarisation calibration. Instead, we directly solve for D and X terms.
Set the polarisation model
First, set the polarisation model for the polarised position-angle calibrator (here 1331+305=3C286 which is also our primary flux calibrator). For polarisation properties of your primary polarisation calibrator see VLA Polarimetry Guide.
i0=7.3109 # Stokes I value for spw 0 ch 0
f0=4.8851 # Frequency for spw0 ch0 (note that in our data the 'lower' spw is actually higher frequency)
alpha=log(i0/7.35974932)/log(4.8351/f0) # Values from our setjy() run on Stokes I earlier
c0=0.114 # Fractional polarisation 11.4% for 5GHz
d0=33*pi/180 # Polarisation angle 33deg in radians
myPolSetjy = setjy(vis='jupiter6cm.demo.ms', field='1331+305', standard='manual', spw='', fluxdensity=[i0,0,0,0], spix=[alpha,0], reffreq=str(f0)+'GHz', polindex=[c0,0], polangle=[d0,0], scalebychan=True, usescratch=False)
The results are displayed in the CASA logger as well as saved in the myPolSetjy python dictionary
#In CASA
CASA <49>: myPolSetjy
Out[49]:
{'7': {'0': {'fluxd': array([ 7.3109 , 0.33899165, 0.7613877 , 0. ])},
'1': {'fluxd': array([ 7.26237491, 0.33674164, 0.7563341 , 0. ])},
'fieldName': '1331+305'},
'format': "{field Id: {spw Id: {fluxd: [I,Q,U,V] in Jy}, 'fieldName':field name }}"}
Solve for the leakage terms (D terms)
Solve for polarization leakage on 0137+331. In this step we solve for the instrumental polarization. We will assume here our calibrator has unknown polarisation (poltype='D+QU' if good parallactic coverage, 'D' otherwise, consult polcal() help for more information on options).
polcal(vis='jupiter6cm.demo.ms', caltable='jupiter6cm.demo.D', field='0137+331', spw='', refant='11', poltype='D+QU', solint='inf', combine='scan',minsnr=3, gaintable=['jupiter6cm.demo.gc', 'jupiter6cm.demo.G'], gainfield=['','0137+331',''])
Check the solutions
plotms(vis='jupiter6cm.demo.D', xaxis='antenna', yaxis='amp', showgui=True, plotfile='jupiter6cm.demo.D.amp.png')
plotms(vis='jupiter6cm.demo.D', xaxis='antenna', yaxis='phase', plotrange=[-1,-1,-200,200], showgui=True, plotfile='jupiter6cm.demo.D.phase.png')
plotms(vis='jupiter6cm.demo.D', xaxis='antenna', yaxis='snr', showgui=True, plotfile='jupiter6cm.demo.D.snr.png')
Solve for the R-L polarisation angle (X term)
The total polarisation is now correct (since we just calibrated the instrumental polarisation, i.e. D terms), but now the R-L phase needs to be calibrated to obtain an accurate polarisation position angle.
polcal(vis='jupiter6cm.demo.ms', caltable='jupiter6cm.demo.X', field='1331+305', spw='', refant='11', poltype='Xf', solint='inf', combine='scan',minsnr=3, gaintable=['jupiter6cm.demo.gc', 'jupiter6cm.demo.G', 'jupiter6cm.demo.D'], gainfield=['','1331+305','',''])
Check the solutions in CASA logger window
The following calibration term is arranged for solve: . Xf Jones: table=jupiter6cm.demo.X append=false solint=inf,none refantmode='flex' refant='none' minsnr=3 apmode=AP solnorm=false For solint = inf, found 2 solution intervals. Mean position angle offset solution for 1331+305 (spw = 0) = 80.7525 deg. Mean position angle offset solution for 1331+305 (spw = 1) = 48.9853 deg. Found good Xf Jones solutions in 2 solution intervals.
NOTE: If you are using CASA 4.7 or earlier, you may want to use poltype='X' in X-term calculations (Mueller matrices). For CASA 5.0 and later this option will not work (the relevant CASA documentation is being updated), and hence you need to use poltype='Xf' (Jones matrices). Although 'Xf' should be used predominantly for large bandwidths, which clearly is not the case here, it can also be used for the single channel old VLA data as long as you make sure the interp parameter is set to default (which should be nearest).
Apply the calibration
Now that we have derived all the calibration solutions and saved them in the tables of multiple extensions, we need to apply them to the actual data. The following tasks will apply the solution tables to the DATA and write a new column CORRECTED_DATA as it is standard for CASA. Important: make sure you set parang=True.
applycal(vis='jupiter6cm.demo.ms', field='1331+305', spw='', selectdata=False, gaintable=['jupiter6cm.demo.gc', 'jupiter6cm.demo.G', 'jupiter6cm.demo.D','jupiter6cm.demo.X'], gainfield=['','1331+305'], calwt=[False], parang=True)
applycal(vis='jupiter6cm.demo.ms', field='0137+331', spw='', selectdata=False, gaintable=['jupiter6cm.demo.gc', 'jupiter6cm.demo.G', 'jupiter6cm.demo.Gflx','jupiter6cm.demo.D','jupiter6cm.demo.X'], gainfield=['','0137+331','0137+331'], calwt=[False], parang=True)
applycal(vis='jupiter6cm.demo.ms', field='JUPITER', spw='', selectdata=False, gaintable=['jupiter6cm.demo.gc', 'jupiter6cm.demo.G', 'jupiter6cm.demo.Gflx','jupiter6cm.demo.D','jupiter6cm.demo.X'], gainfield=['','0137+331','0137+331'], calwt=[False], parang=True)
It's a good idea to split the Jupiter target data at this point. We will write the corrected data of our target source to a new single-source MS.
split(vis='jupiter6cm.demo.ms', outputvis='jupiter6cm.demo.JUPITER.split.ms', field='JUPITER', datacolumn='corrected')
You can use plotms() task to look at the split calibrated data.
Initial imaging (Stokes I)
We will now image and clean the calibrated Jupiter data. In the next step we will perform self-calibration, so here in the initial imaging we will not clean too deeply and we will also save the model in the MS data (with the use of parameter savemodel in tclean()).
Since we set mask="" in our cleaning tasks, the interactive GUI will not allow you to do the deconvolution before you draw the mask on at least one plane. When drawing a mask, make sure to extend it to all channels and all polarisations.
tclean(vis='jupiter6cm.demo.JUPITER.split.ms', stokes='I', field='', spw='', imagename='jupiter6cm.demo.JUPITER.I.clean1', robust = 0.0, imsize=[288], cell=['3arcsec'], specmode='mfs', deconvolver='hogbom', weighting='briggs', threshold='0.1mJy', mask='', niter=500, interactive=True, cycleniter=100, savemodel='modelcolumn', pblimit=-0.2)
Inspect the quality of the cleaned images with imstat().
imstat(imagename='jupiter6cm.demo.JUPITER.I.clean1.image', stokes='I', box='216,1,287,72')
In this initial imaging the deconvolution will stop with the iteration limit since we set it fairly low. We 'cleaned' approximately 4.3 Jy in this initial imaging step, and reached rms of about 3.52 mJy/bm.
Self-calibration
In the self calibration step we run gaincal() that will make use of the newly created MODEL column in our MS file.
gaincal(vis='jupiter6cm.demo.JUPITER.split.ms', caltable='jupiter6cm.demo.JUPITER.split.selfcal1', field='', spw='', gaintype='G', calmode='p', solint='30s', combine='', refant='11', parang=False, append=False, selectdata=False)
Inspect the solutions
plotms(vis='jupiter6cm.demo.JUPITER.split.selfcal1',xaxis='time',yaxis='amp',gridrows=2, gridcols=3, iteraxis='antenna')
plotms(vis='jupiter6cm.demo.JUPITER.split.selfcal1',xaxis='time',yaxis='phase', gridrows=2, gridcols=3 ,iteraxis='antenna',plotrange=[-1,-1,-180,180])
And apply the calibration to the DATA(creating a CORRECTED column in the MS)
applycal(vis='jupiter6cm.demo.JUPITER.split.ms', field='', spw='', selectdata=False, gaintable=['jupiter6cm.demo.JUPITER.split.selfcal1'], gainfield=[''], interp=['nearest'],calwt=[False], applymode='calflag')
Now, image the self-calibrated data
tclean(vis='jupiter6cm.demo.JUPITER.split.ms', stokes='I', field='', spw='', imagename='jupiter6cm.demo.JUPITER.I.clean2', robust= 0.0, imsize=[288], cell=['3arcsec'], specmode='mfs', deconvolver='hogbom', weighting='briggs', threshold='0.05mJy', mask='', niter=10000, interactive=True, cycleniter=100, pblimit=-0.2)
Check the image statistics
imstat(imagename='jupiter6cm.demo.JUPITER.I.clean2.image', stokes='I', box='216,1,287,72')
After one cycle of self-calibration with tclean() we reached rms=1.10 mJy/bm. But bear in mind that the rms values may differ for you since they will depend on how deeply you cleaned and how good a mask you applied.
Full Stokes Imaging
At this stage we will perform full Stokes imaging to create image of each Stokes parameter that we will later use to create polarised intensity and angle images.
tclean(vis='jupiter6cm.demo.JUPITER.split.ms', stokes='IQUV', field='', spw='', imagename='jupiter6cm.demo.JUPITER.IQUV.clean', robust= 0.0, imsize=[288], cell=['3arcsec'], specmode='mfs', deconvolver='clarkstokes', weighting='briggs', threshold='0.05mJy', mask='', niter=10000, interactive=True, cycleniter=100, pblimit=-0.2)
Inspect the quality of the cleaned images with imstat() (do it for each Stokes separately, as otherwise imstat() will return an average of all Stokes). You should be reaching values of about 1.1 mJy/bm (I), 0.62 mJy/bm (Q), 0.90 mJy/bm (U), 0.16 mJy/bm (V).
Polarisation Intensity and Position Angle Images
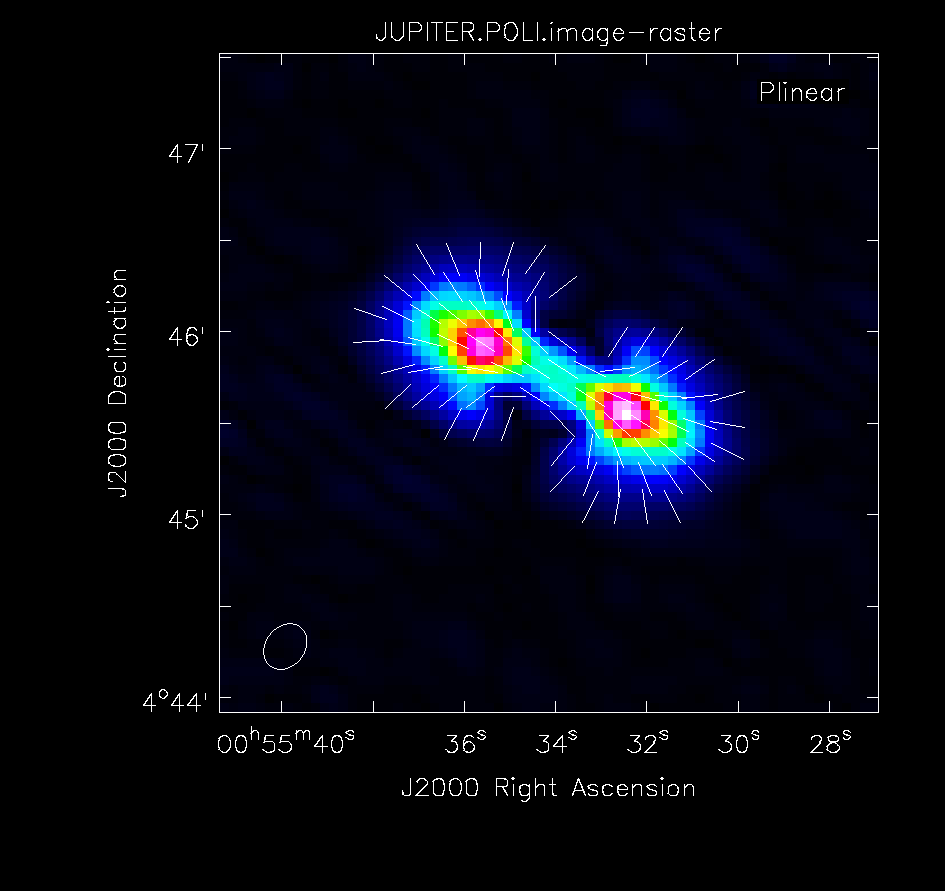
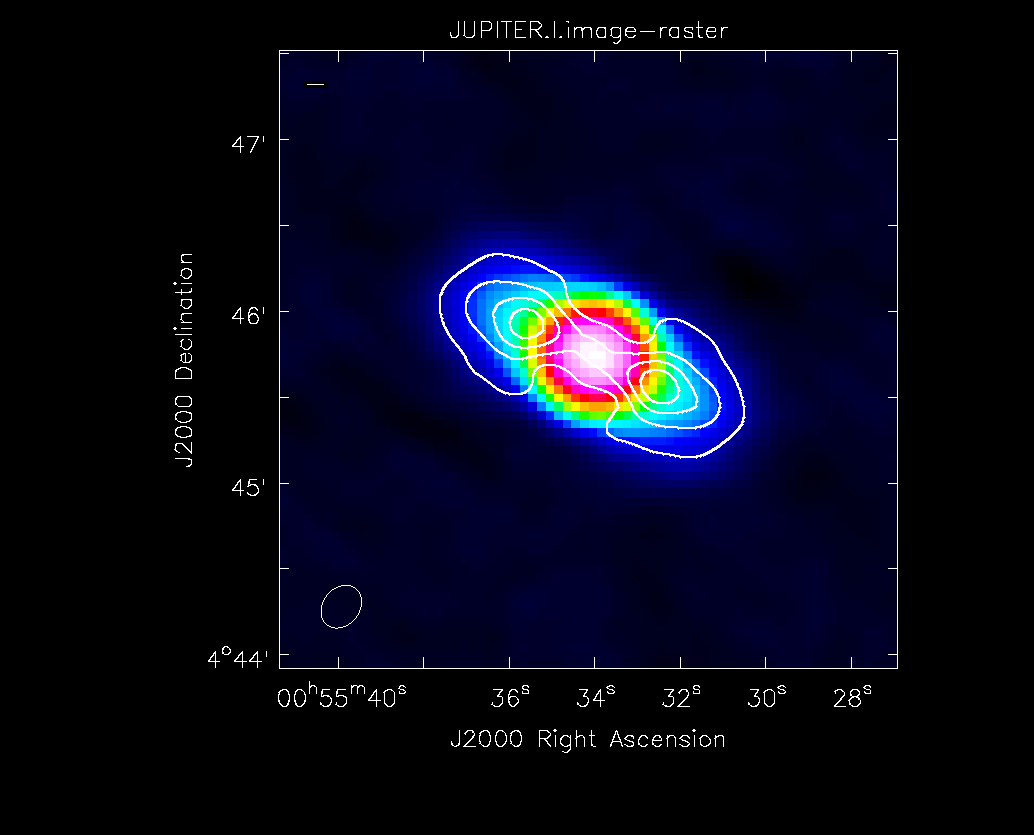
We now want to save each Stokes plane as separate images. We can use either the imsubimage() or immath() tasks to do so.
Run
imsubimage(imagename='jupiter6cm.demo.JUPITER.IQUV.clean.image',outfile='JUPITER.I.image',stokes='I')
imsubimage(imagename='jupiter6cm.demo.JUPITER.IQUV.clean.image',outfile='JUPITER.Q.image',stokes='Q')
imsubimage(imagename='jupiter6cm.demo.JUPITER.IQUV.clean.image',outfile='JUPITER.U.image',stokes='U')
or
immath(imagename='jupiter6cm.demo.JUPITER.IQUV.clean.image', mode='evalexpr', stokes='I', expr='IM0', outfile='JUPITER.I.image')
immath(imagename='jupiter6cm.demo.JUPITER.IQUV.clean.image', mode='evalexpr', stokes='Q', expr='IM0', outfile='JUPITER.Q.image')
immath(imagename='jupiter6cm.demo.JUPITER.IQUV.clean.image', mode='evalexpr', stokes='U', expr='IM0', outfile='JUPITER.U.image')
Finally, we want to create the polarization intensity (POLI) and position angle (POLA) images.
immath(imagename=['JUPITER.Q.image','JUPITER.U.image'], mode='poli', outfile='JUPITER.POLI.image')
We will now use the polithresh parameter when creating the polarisation angle image to get rid of the noise. First check the statistic in the POLI image, and then use it as a threshold while creating the POLA image (in the example below equal to 4 sigma).
imstat(imagename='JUPITER.POLI.image', box='216,1,287,72')
immath(imagename=['JUPITER.Q.image','JUPITER.U.image'], mode='pola', outfile='JUPITER.POLA.image',polithresh='4.5mJy/beam')
Again, check the statistics of these final images with imstat() and/or viewer().
And that's it. For the tutorial on how to perform analysis, display and manipulation of the polarisation images now, consult the appropriate VLA CASA guides.