M100 Band3 SingleDish 4.2.2: Difference between revisions
m Cbrogan moved page M100 Band3 SingleDish 4.1 to M100 Band3 SingleDish 4.2.2: Delay in finishing requires new CASA version |
|||
| (103 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<div style="background-color:#E0FFFF;border:4px solid #FF9966;"> | |||
<div style="font-size:250%; color:red; text-align:center;"> | |||
This page is currently under construction. | |||
</div> | |||
<div style="font-size:200%; color:black; text-align:center"> | |||
DO NOT USE IT. | |||
</div> | |||
<div style="font-size:150%; color:black; text-align:center"> | |||
To navigate the CASAguides pages, visit [http://casaguides.nrao.edu/ | |||
casaguides.nrao.edu | |||
] | |||
</div> | |||
</div> | |||
M100 Single Dish Data Reduction (under modification by AH and CV) | |||
[[Category:ALMA]][[Category:Calibration]][[Category:Spectral Line]] | |||
*'''This guide requires CASA 4.1.0 and assumes that you have downloaded M100_Band3_SD_UncalibratedData.tgz from [[M100_Band3#Obtaining_the_Data]]''' | |||
*'''Details of the ALMA observations are provided at [[M100_Band3]] | |||
*'''This portion of the guide covers calibration of the raw visibility data. To skip to the imaging portion of the guide, see: [[M100_Band3_Combine_4.1]]'''. | |||
==Overview== | ==Overview== | ||
This portion of the '''[[ | This portion of the '''[[M100_Band3_SingleDish_4.1]]''' CASA Guide will cover the reduction of the Total Power (TP) array data into units of Kelvins on the antenna temperature (Ta*) scale and imaging. | ||
Converting this image to the Jansky scale (Jy/beam) to be combined with interferometric data is covered in the '''[[M100 Band3 Combine 4.1]]''' section. | |||
'''This guide is designed for CASA 4.1.0. | '''This guide is designed for CASA 4.1.0. | ||
| Line 10: | Line 31: | ||
If you haven't downloaded the data, you can XXXXXXX and XXXXXX: | If you haven't downloaded the data, you can XXXXXXX and XXXXXX: | ||
Once the download has finished, | Once the download has finished, unpack the file: | ||
<source lang="bash"> | <source lang="bash"> | ||
| Line 22: | Line 43: | ||
</source> | </source> | ||
== Summary of | == CASA Version == | ||
This guide has been written for CASA release 4.1.0. Please confirm your version before proceeding. | |||
<source lang="python"> | |||
# In CASA | |||
version = casadef.casa_version | |||
print "You are using " + version | |||
if (version < '4.1.0'): | |||
print "YOUR VERSION OF CASA IS TOO OLD FOR THIS GUIDE." | |||
print "PLEASE UPDATE IT BEFORE PROCEEDING." | |||
else: | |||
print "Your version of CASA is appropriate for this guide." | |||
</source> | |||
== Summary of Datasets == | |||
There were six observations made. The table below indicates the uid reference of each and the start and end times. | There were six observations made. The table below indicates the uid reference of each and the start and end times. | ||
| Line 35: | Line 71: | ||
</pre> | </pre> | ||
We will eventually concatenate the six datasets used here into one large dataset. However, we will keep them separate for now, as some of the steps to follow require individual datasets to be calibrated separately (namely, the sky/Tsys calibration and baseline subtraction). We therefore start by defining an array "basename" that includes the names of the six files in chronological order. This will simplify the following steps by allowing us to loop through the files using a simple for-loop in python. Remember that if you log out of CASA, you will have to re-issue this command. We will remind you of this in the relevant sections by repeating the command at the start. | We will eventually concatenate the six datasets used here into one large dataset. However, we will keep them separate for now, as some of the steps to follow require individual datasets to be calibrated separately (namely, the sky/Tsys calibration and baseline subtraction). We therefore start by defining an array "basename" that includes the names of the six files in chronological order. This will simplify the following steps by allowing us to loop through the files using a simple for-loop in python. Remember that if you log out of CASA, you will have to re-issue this command. We will remind you of this in the relevant sections by repeating the command at the start. | ||
| Line 59: | Line 78: | ||
</source> | </source> | ||
== | == Create Measurement Sets == | ||
The raw data have been provided to you in the ASDM format. ASDM stands for ALMA Science Data Model. It is the native format of the data produced by the observatory. Before we can proceed to the calibration, we will need to convert those data to the CASA MS format. This is done simply with the task importasdm. | The raw data have been provided to you in the ASDM format. ASDM stands for ALMA Science Data Model. It is the native format of the data produced by the observatory. | ||
Before we can proceed to the calibration, we will need to convert those data to the CASA MS format. This is done simply with the task importasdm. | |||
<source lang="python"> | <source lang="python"> | ||
In CASA | #In CASA | ||
importasdm( | for name in basename: | ||
importasdm(asdm = name) | |||
</source> | </source> | ||
Note: importasdm has an option singledish, which you may be tempted to use. It works, but it has some limitations (which will be removed in the future), so for now, we recommend not using it. | Note: importasdm has an option singledish, which you may be tempted to use. It works, but it has some limitations (which will be removed in the future), so for now, we recommend not using it. | ||
== Initial Inspection == | |||
The usual first step is then to get some basic information about the data. We do this using the task {{listobs}}, which will output a detailed summary of each dataset supplied. | The usual first step is then to get some basic information about the data. We do this using the task {{listobs}}, which will output a detailed summary of each dataset supplied. | ||
| Line 80: | Line 104: | ||
The output will be sent to the CASA [http://casa.nrao.edu/docs/userman/UserMansu41.html#UserMansu42.html logger]. | The output will be sent to the CASA [http://casa.nrao.edu/docs/userman/UserMansu41.html#UserMansu42.html logger]. | ||
You will have to scroll up to see the individual output for each of the six datasets. Here is an example of the most relevant output for the fifth file in the list. | You will have to scroll up to see the individual output for each of the six datasets. Here is an example of the most relevant output for the fifth file in the list. | ||
These commands define a python list called "basename", which contains the name of all 6 MS files. | |||
The "for" loop executes for each element in basename, calling listobs and directing the output to a file called, e.g., "uid___A002_X60b415_X39a.ms.listobs.txt" for the first measurement set. | |||
You can browse through the listobs output as you would normally look at a text file (use emacs, vi, or another editor). You can also send the output to the terminal from inside of CASA. To do so type: | |||
<source lang="python"> | |||
# In CASA | |||
os.system('cat uid___A002_X60b415_X39a.ms.listobs.txt') | |||
</source> | |||
or | |||
<source lang="python"> | |||
# In CASA | |||
os.system('more uid___A002_X60b415_X39a.ms.listobs.txt') | |||
</source> | |||
CASA knows a few basic shell commands like 'cat', 'ls', and 'rm' but for more complex commands you will need to run them inside 'os.system("command")'. For more information see http://casa.nrao.edu/ . | |||
Here is an example of the (abridged) output from {{listobs}} for the first dataset in the list, uid___A002_X60b415_X39a.ms. You would see this if you had specified '''verbose''' to be False in the listobs call: | |||
<pre style="background-color: #fffacd;"> | <pre style="background-color: #fffacd;"> | ||
| Line 164: | Line 208: | ||
</pre> | </pre> | ||
This output shows that three sources were observed in each data set: M100. | |||
* '''M100''' are our science target. Note that the source corresponds to a number of individual fields (see the Field ID column). | |||
The output also shows that the data contain many spectral windows. Using the labeling scheme in the listobs above these are: | |||
* '''spw 9''','''spw 11''','''spw 13''' and '''spw 15''' hold our science data. These are "Frequency Domain Mode" (FDM) data with small (0.49 MHz) channel width and wide total bandwidth. As a result these have a lot of channels (4080). spw 15 holds the lower sideband (LSB) data and includes the CO(1-0) line. | |||
We will focus on these data. | |||
* '''spw 1''','''spw 3''','''spw 5''' and '''spw 7''' hold lower a resolution processing ("Time Domain Mode", TDM) of the data from the same part of the spectrum (baseband). These data have only 128 channels across 2 GHz bandwidth and so have a much coarser channel spacing than the FDM data. | |||
These were used to generate the calibration tables that we include in the tarball but will not otherwise appear in this guide. | |||
We do some initial inspection of the data using plotms. First we will plot amplitude versus channel, averaging over time in order to speed up the plotting process. | |||
<source lang="python"> | |||
# In CASA | |||
plotms(vis='uid___A002_X60b415_X39a.ms', xaxis='channel', yaxis='amp', | |||
averagedata=T, avgtime='1e8', avgscan=T, iteraxis='antenna') | |||
</source> | |||
Next, we will look at amplitude versus time, averaging over channels and colorizing by field. | |||
<source lang="python"> | |||
# In CASA | |||
plotms(vis='uid___A002_X60b415_X39a.ms', xaxis='time', yaxis='amp', | |||
averagedata=T, avgchannel='100', iteraxis='spw', coloraxis='field') | |||
</source> | |||
== Convert MS format to single-dish format == | |||
In order to calibrate the data we need the data to be in the single-dish ASAP format. Most of the tasks that we will use for calibration are part of the ASAP package, which has been incorporated into CASA. The ASAP package uses a different data format, so from a global point of view, what we are going to do is, first convert the MS to the ASAP format, then run the necessary calibration tasks, then convert the data back to the MS format. Another important difference with interferometric data reduction is that the calibration is performed directly on the dataset, we will not produce calibration tables and apply them at the end. An effort is on-going to update the SD routines so that this is done, that should be available soon, but until then, please remember that all SD calibration operations apply to the data directly, so you may want to always create a new dataset each time, so that you do not have to start all over again. | |||
We are now going to go through each of them with a bit more explanation. We will take the example of uid___A002_X6218fb_X264.ms. | |||
We convert the data to the ASAP format using the routine sd.splitant for that. An outprefix has to be specified because the routine will create a dataset per antenna, taking the outprefix, and appending the antenna name and the format extension ('.asap'). | |||
<source lang="python"> | |||
# In CASA | |||
sd.splitant(filename = 'uid___A002_X6218fb_X264.ms', | |||
outprefix = 'uid___A002_X6218fb_X264.ms', | |||
overwrite = True, | |||
freq_tolsr = True) | |||
</source> | |||
The reason for the option freq_tolsr=True is to convert the frequencies in the MS from TOPO to LSRK. This is an important setting, as due to current limitations, it is the only stage where this conversion can be done. Using freq_tolsr=False would keep the frame of the frequencies from the MS, and would force you to image the final cube in TOPO velocities. Another important aspect is for the baselining/line finding: if your observations have been obtained at different epochs, it may be easier to work on spectra with LSRK frequencies because then the line emissions of all spectra can be overlapped. | |||
We have now two ASAP datasets, one for PM03 and one for PM04. As usual, we will first obtain information about the content of the datasets, using sdlist (equivalent of listobs). | |||
<source lang="python"> | |||
# In CASA | |||
sdlist(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.sdlist') | |||
sdlist(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.sdlist') | |||
</source> | |||
Here is an example of the most relevant output for uid___A002_X6218fb_X264.ms.PM04.asap.sdlist. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('more uid___A002_X6218fb_X264.ms.PM04.asap.sdlist') | |||
</source> | |||
<pre style="background-color: #fffacd;"> | |||
-------------------------------------------------------------------------------- | |||
Scan Table Summary | |||
-------------------------------------------------------------------------------- | |||
Project: uid://A002/X5d9e5c/X42 | |||
Obs Date: 2013/04/28/04:10:19 | |||
Observer: cvlahakis | |||
Antenna Name: ALMA//PM04@T703 | |||
Data Records: 15186 rows | |||
Obs. Type: CALIBRATE_ATMOSPHERE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE | |||
Beams: 1 | |||
IFs: 17 | |||
Polarisations: 2 (linear) | |||
Channels: 4080 | |||
Flux Unit: K | |||
Abscissa: Channel | |||
Selection: none | |||
Scan Source Time range Int[s] Record SrcType FreqIDs MolIDs | |||
Beam Position (J2000) | |||
-------------------------------------------------------------------------------- | |||
0 M100 2013/04/28/04:12:06.1 - 04:13:17.3 0.500364 594 [PSON:CALON, PSOFF:CALON] [0, 1, 2, 3, 4, 5, 6, 7, 8] [0] | |||
0 J2000 12:23:07.0 +15.51.15.9 | |||
1 M100 2013/04/28/04:14:05.0 - 04:15:50.8 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.9 +15.51.17.4 | |||
2 M100 2013/04/28/04:16:09.4 - 04:17:56.4 1.01628 1443 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.8 +15.51.19.1 | |||
3 M100 2013/04/28/04:18:13.8 - 04:20:00.8 1.01628 1443 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.8 +15.51.20.6 | |||
4 M100 2013/04/28/04:20:19.4 - 04:22:05.2 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.7 +15.51.22.2 | |||
5 M100 2013/04/28/04:22:23.8 - 04:24:09.7 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.6 +15.51.23.7 | |||
6 M100 2013/04/28/04:25:03.7 - 04:26:14.7 0.500364 594 [PSON:CALON, PSOFF:CALON] [0, 1, 2, 3, 4, 5, 6, 7, 8] [0] | |||
0 J2000 12:23:06.5 +15.51.25.6 | |||
7 M100 2013/04/28/04:26:33.8 - 04:28:19.6 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.5 +15.51.26.7 | |||
8 M100 2013/04/28/04:28:38.2 - 04:30:25.2 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.4 +15.51.28.1 | |||
9 M100 2013/04/28/04:30:42.6 - 04:32:29.6 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.3 +15.51.29.5 | |||
10 M100 2013/04/28/04:32:48.2 - 04:34:34.0 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.2 +15.51.30.9 | |||
11 M100 2013/04/28/04:34:52.6 - 04:36:08.5 1.0162 1018 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4] | |||
0 J2000 12:23:06.2 +15.51.32.3 | |||
-------------------------------------------------------------------------------- | |||
FREQUENCIES: 9 | |||
ID IFNO Frame RefVal RefPix Increment Channels POLNOs | |||
0 0 LSRK 1.87675e+11 1.5 2.5e+09 4 [0] | |||
1 1 LSRK 1.0095e+11 63.5 -15625000 128 [0, 1] | |||
2 2 LSRK 1.0092656e+11 0 -1.78125e+09 1 [0, 1] | |||
3 3 LSRK 1.0276515e+11 63.5 -15625000 128 [0, 1] | |||
4 4 LSRK 1.0274171e+11 0 -1.78125e+09 1 [0, 1] | |||
5 5 LSRK 1.1280715e+11 63.5 15625000 128 [0, 1] | |||
6 6 LSRK 1.1278371e+11 0 1.78125e+09 1 [0, 1] | |||
7 7 LSRK 1.1468215e+11 63.5 15625000 128 [0, 1] | |||
8 8 LSRK 1.1465871e+11 0 1.78125e+09 1 [0, 1] | |||
9 9 LSRK 1.0095e+11 2039.5 -488281.25 4080 [0, 1] | |||
10 10 LSRK 1.0094976e+11 0 -1.9921875e+09 1 [0, 1] | |||
11 11 LSRK 1.0276515e+11 2039.5 -488281.25 4080 [0, 1] | |||
12 12 LSRK 1.0276491e+11 0 -1.9921875e+09 1 [0, 1] | |||
13 13 LSRK 1.1280715e+11 2039.5 488281.25 4080 [0, 1] | |||
14 14 LSRK 1.1280691e+11 0 1.9921875e+09 1 [0, 1] | |||
15 15 LSRK 1.1468215e+11 2039.5 488281.25 4080 [0, 1] | |||
16 16 LSRK 1.1468191e+11 0 1.9921875e+09 1 [0, 1] | |||
-------------------------------------------------------------------------------- | |||
MOLECULES: | |||
ID RestFreq Name | |||
0 [] [] | |||
1 [1.0095e+11] [Manual_window(ID=0)] | |||
2 [1.02794e+11] [Manual_window(ID=0)] | |||
3 [1.12794e+11] [Manual_window(ID=0)] | |||
4 [1.14669e+11] [CO_v_0_1_0(ID=3768098)] | |||
-------------------------------------------------------------------------------- | |||
</pre> | |||
== Calibration == | == Calibration == | ||
About the calibration itself: the two main steps are | About the calibration itself: the two main steps are | ||
== | <pre style="background-color: #fffacd;"> | ||
1. Calibration of the spectra into K, by applying the Tsys calibration and removing the signal from the OFF position | |||
2. Baselines Subtraction (i.e. subtracting the background emission, to keep only the line emission.) | |||
</pre> | |||
=== Tsys Correction === | |||
Let's start by checking the Tsys solutions. We will use the gencal command used for interferometric data reduction. | |||
This will produce a CASA calibration table, which can be analysed similarly to previous guides '''[[M100 Band3 ACA 4.1#Tsys]]''' | |||
<source lang="python"> | |||
# In CASA | |||
gencal(vis = 'uid___A002_X6218fb_X264.ms', | |||
caltable = 'uid___A002_X6218fb_X264.ms.tsys', | |||
caltype = 'tsys') | |||
plotbandpass(caltable='uid___A002_X6218fb_X264.ms.tsys', overlay='time', | |||
xaxis='freq', yaxis='amp', subplot=22, buildpdf=False, interactive=False, | |||
showatm=True,pwv='auto',chanrange='5~123',showfdm=True, | |||
field='', figfile='uid___A002_X6218fb_X264.ms.tsys.plots.overlayTime/uid___A002_X6218fb_X264.ms.tsys') | |||
</source> | |||
This sequence loops over all of our files and plots Tsys as a function of time for channel. In the call to {{plotcal}}: | |||
* '''subplot'''=22 parameter sets up a 2 x 2 panel grid. | |||
* '''iteration''' tells {{plotcal}} to make a separate plot for each antenna. | |||
<figure id="X264.Tsys.png"> | |||
[[File:X264.Tsys.png|200px|thumb|right|<caption> Tsys vs. frequency plot for uid___A002_X6218fb_X264.</caption>]] | |||
</figure> | |||
==== Fill Tsys Column ==== | |||
For the spectral window association, we will use the routine tsysspwmap available directly in CASA. | |||
<source lang="python"> | |||
# In CASA | |||
from recipes.almahelpers import tsysspwmap | |||
tsysmap = tsysspwmap(vis = 'uid___A002_X6218fb_X264.ms', tsystable = 'uid___A002_X6218fb_X264.ms.tsys') | |||
</source> | |||
tsysmap is a list of indices, with tsysmap[i] being the index of the Tsys spw to associate to the science spw of index i. | |||
In the next step, we will do the actual preparation of the Tsys (i.e. re-sampling and re-indexing of the Tsys spw), using the library filltsys, also available directly in CASA. | |||
<source lang="python"> | <source lang="python"> | ||
In CASA | # In CASA | ||
import filltsys | |||
for i in [9, 11, 13, 15]: | |||
filltsys.fillTsys('uid___A002_X6218fb_X264.ms.PM03.asap', | |||
specif = i, | |||
tsysif = tsysmap[i], | |||
mode = 'linear', | |||
extrap = True) | |||
for i in [9, 11, 13, 15]: | |||
filltsys.fillTsys('uid___A002_X6218fb_X264.ms.PM04.asap', | |||
specif = i, | |||
tsysif = tsysmap[i], | |||
mode = 'linear', | |||
extrap = True) | |||
</source> | |||
This will loop through each science spw, re-sample the Tsys solutions associated to spw of index tsysmap[i] using a linear interpolation, allowing for extrapolation in case the science and Tsys spw do not perfectly match, | |||
and re-index it to spw of index i. | |||
'''You will note that the SD tasks use the word 'if': this is equivalent to 'spw id' in interferometry tasks.''' | |||
==== View Spectra ==== | |||
<source lang="python"> | |||
# In CASA | |||
sdplot(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap', | |||
iflist = [15], | |||
scanaverage = True, | |||
plottype = 'spectra', | |||
stack = 'pol', | |||
panel = 'scan', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.png', | |||
overwrite = True) | |||
</source> | |||
=== A Priori Flagging === | |||
We have now prepared the Tsys solutions for application. We have not applied them yet. We will first do some a-priori flagging, of the edge channels. This is similar to flagging edge channels in TDM interferometry datasets. In FDM interferometry datasets, these edge channels are not written out, so there is no need to flag anything. Here, we have an FDM SD dataset, produced with the second correlator (the ACA correlator), which does not excise them (yet). | |||
Each spw covers the full baseband width (2GHz), so we will flag 120 channels on each side so as to keep only the center 3080 channels, similarly to FDM interferometry datasets. | |||
<source lang="python"> | |||
# In CASA | |||
sdflag(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap', | |||
specunit = 'channel', | |||
iflist = [9, 11, 13, 15], | |||
maskflag = [[0, 119], [3960, 4079]], | |||
overwrite = True) | |||
sdflag(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap', | |||
specunit = 'channel', | |||
iflist = [9, 11, 13, 15], | |||
maskflag = [[0, 119], [3960, 4079]], | |||
overwrite = True) | |||
</source> | |||
If you want to flag data in tool base, we will present an example for that. | |||
<source lang="python"> | |||
# In CASA | |||
param_org = sd.rcParams['scantable.storage'] | |||
sd.rcParams['scantable.storage'] = 'disk' | |||
s = sd.scantable('uid___A002_X6218fb_X264.ms.PM03.asap', average=False) | |||
sel = sd.selector() | |||
sel.set_ifs([9, 11, 13, 15]) | |||
s.set_selection(sel) | |||
mask = s.create_mask([[0, 119], [3960, 4079]]) | |||
s.flag(mask) | |||
s.set_selection() | |||
del s | |||
sd.rcParams['scantable.storage'] = param_org | |||
</source> | |||
=== Apply Calibration and Inspect === | |||
Now is the step where we will actually calibrate the data. This is done with the task sdcal. This is a straight-forward operation, so no opportunity for tweaking. The calibration mode (calmode) shall be 'ps', as in Position Switching. The task will find automatically the OFF-position measurements. We need to specify the science spws (iflist) because they are the only ones that can be calibrated. We specify a different output file (outfile) so as to be able to come back to the original dataset if necessary. | |||
<source lang="python"> | |||
# In CASA | |||
sdcal(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap', | |||
calmode = 'ps', | |||
iflist = [9, 11, 13, 15], | |||
scanaverage = False, | |||
timeaverage = False, | |||
polaverage = False, | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal', | |||
overwrite = True) | |||
sdcal(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap', | |||
calmode = 'ps', | |||
iflist = [9, 11, 13, 15], | |||
scanaverage = False, | |||
timeaverage = False, | |||
polaverage = False, | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal', | |||
overwrite = True) | |||
</source> | |||
Before proceeding to the next step, we can plot the calibrated spectra, using the sdplot task. The commands below will plot one spectrum per scan, spw and polarization. It is difficult to describe what a good spectrum looks like at this stage. What you should look for is outliers (i.e. spectra that differ from most, whether in shape or level). | |||
<source lang="python"> | |||
# In CASA | |||
for i in [9, 11, 13, 15]: | |||
sdplot(infile='uid___A002_X6218fb_X264.ms.PM03.asap.cal', | |||
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan', | |||
outfile='uid___A002_X6218fb_X264.ms.PM03.asap.cal.plots/uid___A002_X6218fb_X264.ms.PM03.asap.cal.spectra.spw'+str(i)+'.png', | |||
overwrite = True) | |||
for i in [9, 11, 13, 15]: | |||
sdplot(infile='uid___A002_X6218fb_X264.ms.PM04.asap.cal', | |||
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan', | |||
outfile='uid___A002_X6218fb_X264.ms.PM04.asap.cal.plots/uid___A002_X6218fb_X264.ms.PM04.asap.cal.spectra.spw'+str(i)+'.png', | |||
overwrite = True) | |||
</source> | |||
==== View Calibrated Spectra ==== | |||
<source lang="python"> | |||
# In CASA | |||
sdplot(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal', | |||
iflist = [15], | |||
scanaverage = True, | |||
plottype = 'spectra', | |||
stack = 'pol', | |||
panel = 'scan', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.png', | |||
overwrite = True) | |||
</source> | |||
<figure id="X264.PM04.cal.png"> | |||
[[File:X264.PM04.cal.png|200px|thumb|right|<caption> Brightness temperature vs. channel plot after calibration for M100 scans.</caption>]] | |||
</figure> | |||
=== Baseline Subtraction and Inspect === | |||
We will now perform the second main step of the calibration: the baselining. This is done with the sdbaseline task. | |||
In the commands below, the baseline fitting method is controlled by the parameters blfunc and order. Many fitting methods are available (’poly’,’chebyshev’,’cspline’,’sinusoid’, see help of sdbaseline task for more information). Here, we will do a simple linear fitting. | |||
The parameter maskmode allows excluding portions of the spectra from the baseline fitting. This is mainly useful to exclude channels where there are line emissions (it could also be used to excluse the edge channels, in case they were not flagged beforehand). The channels can be specified as a list or chosen interactively. Another option is to use the line finder implemented in the sdbaseline task, using maskmode = 'auto'; then the parameters thresh and avg_limit can be used to tweak the line finding algorithm. (thresh is specified in sigma units, avg_limit is specified as a number of channels.) | |||
<source lang="python"> | |||
# In CASA | |||
sdbaseline(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal', | |||
iflist = [9, 11, 13, 15], | |||
maskmode = 'auto', | |||
thresh = 3.0, | |||
avg_limit = 8, | |||
blfunc = 'poly', | |||
order = 1, | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl', | |||
overwrite = True) | |||
sdbaseline(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal', | |||
iflist = [9, 11, 13, 15], | |||
maskmode = 'auto', | |||
thresh = 3.0, | |||
avg_limit = 8, | |||
blfunc = 'poly', | |||
order = 1, | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl', | |||
overwrite = True) | |||
</source> | |||
We run again sdplot to check the output spectra, fully calibrated this time. | |||
<source lang="python"> | |||
# In CASA | |||
for i in [9, 11, 13, 15]: | |||
sdplot(infile='uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl', | |||
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan', | |||
outfile='uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.plots/uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.spectra.spw'+str(i)+'.png', | |||
overwrite = True) | |||
for i in [9, 11, 13, 15]: | |||
sdplot(infile='uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl', | |||
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan', | |||
outfile='uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl.plots/uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl.spectra.spw'+str(i)+'.png', | |||
overwrite = True) | |||
</source> | |||
'''We have now entirely calibrated the dataset Into Kelvin units. The conversion to Jy will be covered into the guide on combination.''' | |||
==== View Baselined Spectra ==== | |||
<source lang="python"> | |||
# In CASA | |||
sdplot(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl', | |||
iflist = [15], | |||
specunit = 'GHz', | |||
scanaverage = True, | |||
stack = 'pol', | |||
panel = 'scan', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.png', | |||
overwrite = True) | |||
</source> | |||
<figure id="X264.PM04.cal.bl.png"> | |||
[[File:X264.PM04.cal.bl.png|200px|thumb|right|<caption> Brightness temperature vs. channel plot after baseline subtraction for M100 scans.</caption>]] | |||
</figure> | |||
== Prepare for Imaging== | |||
=== Convert each ASAP dataset to an MS === | |||
Here, we will convert all of the calibrated asap dataset into measurement sets. The CASA task {{sdsave}} will do this. | |||
<source lang="python"> | |||
#In CASA | |||
sdsave(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.ms', | |||
outform = 'MS2') | |||
sdsave(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl', | |||
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl.ms', | |||
outform = 'MS2') | |||
</source> | |||
=== Combine all executions to one MS === | |||
Concatenate all of the calibrated measurement sets into one for imaging. The CASA task {{concat}} will do this. | |||
<source lang="python"> | |||
#In CASA | |||
os.system('rm -rf concat_m100.ms') | os.system('rm -rf concat_m100.ms') | ||
concat(vis='uid___A002_X60b415_X39a.ms.cal.split', 'uid___A002_X60b415_X6f7.ms.cal.split', 'uid___A002_X6218fb_X264.ms.cal.split', 'uid___A002_X6218fb_X425.ms.cal.split', 'uid___A002_X6321c5_X3a7.ms.cal.split', 'uid___A002_X6321c5_X5ca.ms.cal.split'], | concat(vis='uid___A002_X60b415_X39a.ms.cal.split', 'uid___A002_X60b415_X6f7.ms.cal.split', 'uid___A002_X6218fb_X264.ms.cal.split', 'uid___A002_X6218fb_X425.ms.cal.split', 'uid___A002_X6321c5_X3a7.ms.cal.split', 'uid___A002_X6321c5_X5ca.ms.cal.split'], | ||
| Line 190: | Line 631: | ||
== Image the Total Power Data == | == Image the Total Power Data == | ||
Run listobs on the total power data to see what spw contains the CO | Run listobs on the total power data to see what spw contains the CO(1-0). | ||
<source lang="python"> | <source lang="python"> | ||
In CASA | #In CASA | ||
os.system('rm -rf concat_m100.ms.listobs') | os.system('rm -rf concat_m100.ms.listobs') | ||
listobs(vis='concat_m100.ms',listfile='concat_m100.ms.listobs') | listobs(vis='concat_m100.ms',listfile='concat_m100.ms.listobs') | ||
</source> | </source> | ||
Spectral window SPWID=3 contains the 115.27 GHz line, so we image this window. The task | Spectral window SPWID=3 contains the 115.27 GHz line, so we image this window. | ||
The task {{sdimaging}} will do this. | |||
<source lang="python"> | <source lang="python"> | ||
In CASA | #In CASA | ||
os.system('rm -rf | os.system('rm -rf M100_SD_cube.image') | ||
sdimaging(infile='concat_m100.ms', | sdimaging(infile='concat_m100.ms', | ||
field=0,spw= | field=0,spw=15, | ||
specunit='km/s',restfreq='115.271204GHz', | specunit='km/s',restfreq='115.271204GHz', | ||
dochannelmap=True, | dochannelmap=True, | ||
nchan=70,start=1400,step=5, | nchan=70,start=1400,step=5, | ||
gridfunction='gjinc',imsize=[50,50], | gridfunction='gjinc',imsize=[50,50], | ||
cell=['10arcsec','10arcsec'], | cell=['10arcsec','10arcsec'], | ||
outfile=' | phasecenter = 'J2000 12h22m54.9 +15d49m15' | ||
outfile='M100_SD_cube.image') | |||
</source> | </source> | ||
| Line 216: | Line 659: | ||
Start and step parameters are specified in units that the user chooses for specunit. The numbers | Start and step parameters are specified in units that the user chooses for specunit. The numbers | ||
here are chosen so that the resulting image has the same number of channels, velocity range and | here are chosen so that the resulting image has the same number of channels, velocity range and | ||
channel width as the 7m and 12m array images. The gridfunction is the weighting function that | channel width as the 7m and 12m array images. | ||
is used to grid the observed flux to individual pixels in the image. | |||
which minimizes aliasing effects. "BOX" | <figure id="X264.M100.chan.png"> | ||
of 1 pixel. | [[File:X264.M100.chan.png|200px|thumb|right|<caption> Channel maps for M100.</caption>]] | ||
the effective diameter of an ALMA 12m antenna. | </figure> | ||
defined by additional subparameters (truncate and gwidth). "GJINC" | |||
the Bessel function, and can minimize the broadening of the effective beam. Any of the functions | |||
which require the | ===Gridding Function=== | ||
dataset, and the user can use the default. | The gridfunction is the weighting function that is used to grid the observed flux to individual pixels in the image. | ||
<pre style="background-color: #fffacd;"> | |||
"SF": a spheroidal function, which minimizes aliasing effects. | |||
"BOX": a pillbox function, which defaults to a kernel box size of 1 pixel. | |||
"PB" (primary beam): assumes an Airy disk, corresponding to an antenna with 10.7m diameter, the effective diameter of an ALMA 12m antenna. | |||
"GAUSS": a gaussian, and its size can be defined by additional subparameters (truncate and gwidth). | |||
"GJINC": a gaussian convolved with the Bessel function, and can minimize the broadening of the effective beam. | |||
</pre> | |||
Any of the functions which require the observing frequency for determining the beam size will read the frequency from the dataset, and the user can use the default. | |||
'''The cell size should be chosen so that it is about 1/3 to 1/4 of the FWHM of the effective beam.''' | |||
== Image Analysis : Moment Maps == | |||
Next we will make moment maps for the CO(1-0) emission: Moment 0 is the integrated intensity; Moment 1 is the intensity weighted velocity field; and Moment 2 is the intensity weighted velocity dispersion. | |||
Above we determined the rms noise levels for M100 mosaics in a line-free and a line-bright channel. | |||
We want to limit the channel range of the moment calculations to those channels with significant emission. One good way to do this is to open the cube in the viewer overlaid with 3-sigma contours, with sigma corresponding to the line-free rms. | |||
For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf M100_SD_cube.image.mom*') | |||
immoments(imagename = 'M100_SD_cube.image', | |||
moments = [0], | |||
axis = 'spectral', | |||
chans = '1~24', | |||
outfile = 'M100_SD_cube.image.mom0') | |||
</source> | |||
For higher order moments it is very important to set a conservative flux threshold. Typically something like 6sigma, using sigma from a bright line channel works well. We do this with the '''mask''' parameter in the commands below. When making multiple moments, {{immoments}} appends the appropriate file name suffix to the value of '''outfile'''. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf M100_SD_cube.image.mom*') | |||
immoments(imagename = 'M100_SD_cube.image', | |||
moments = [1], | |||
axis = 'spectral', | |||
chans = '1~24', | |||
includepix = [0.018, 1.0], | |||
outfile = 'M100_SD_cube.image.mom1') | |||
</source> | |||
== Export data as fits == | |||
If you want to analyze the data using another software package it is easy to convert from CASA format to FITS. | |||
<source lang="python"> | |||
# In CASA | |||
os.system('rm -rf M100_SD_*.fits') | |||
exportfits(imagename='M100_SD_cube.image', fitsimage='M100_SD_cube.image.fits') | |||
exportfits(imagename='M100_SD_cube.image.mom0', fitsimage='M100_SD_mom0.fits') | |||
</source> | |||
Although "FITS format" is supposed to be a standard, in fact most packages expect slightly different things from a FITS image. If you are having difficulty, try setting '''velocity=T''' and/or '''dropstokes=T'''. | |||
==Continue on to Combining Images with 7m and 12m dataset== | |||
Now you can continue on to the [[M100_Band3_Combine_4.1]]. | |||
{{Checked 4.1.0}} | |||
Latest revision as of 14:56, 4 August 2014
This page is currently under construction.
DO NOT USE IT.
To navigate the CASAguides pages, visit [http://casaguides.nrao.edu/ casaguides.nrao.edu ]
M100 Single Dish Data Reduction (under modification by AH and CV)
- This guide requires CASA 4.1.0 and assumes that you have downloaded M100_Band3_SD_UncalibratedData.tgz from M100_Band3#Obtaining_the_Data
- Details of the ALMA observations are provided at M100_Band3
- This portion of the guide covers calibration of the raw visibility data. To skip to the imaging portion of the guide, see: M100_Band3_Combine_4.1.
Overview
This portion of the M100_Band3_SingleDish_4.1 CASA Guide will cover the reduction of the Total Power (TP) array data into units of Kelvins on the antenna temperature (Ta*) scale and imaging. Converting this image to the Jansky scale (Jy/beam) to be combined with interferometric data is covered in the M100 Band3 Combine 4.1 section.
This guide is designed for CASA 4.1.0.
If you haven't downloaded the data, you can XXXXXXX and XXXXXX:
Once the download has finished, unpack the file:
# In a terminal outside CASA
tar -xvzf XXXsingledish_datasetXXX_TBD.tgz
cd XXrelevant_directoryXXX
# Start CASA
casapy
CASA Version
This guide has been written for CASA release 4.1.0. Please confirm your version before proceeding.
# In CASA
version = casadef.casa_version
print "You are using " + version
if (version < '4.1.0'):
print "YOUR VERSION OF CASA IS TOO OLD FOR THIS GUIDE."
print "PLEASE UPDATE IT BEFORE PROCEEDING."
else:
print "Your version of CASA is appropriate for this guide."
Summary of Datasets
There were six observations made. The table below indicates the uid reference of each and the start and end times.
uid___A002_X60b415_X39a Observed from 14-Apr-2013/05:34:15.7 to 14-Apr-2013/05:58:23.8 (UTC) uid___A002_X60b415_X6f7 Observed from 14-Apr-2013/06:23:03.0 to 14-Apr-2013/06:47:11.0 (UTC) uid___A002_X6218fb_X264 Observed from 28-Apr-2013/04:12:06.1 to 28-Apr-2013/04:36:07.5 (UTC) uid___A002_X6218fb_X425 Observed from 28-Apr-2013/04:38:56.8 to 28-Apr-2013/05:03:00.3 (UTC) uid___A002_X6321c5_X3a7 Observed from 12-May-2013/02:22:16.9 to 12-May-2013/02:43:59.8 (UTC) uid___A002_X6321c5_X5ca Observed from 12-May-2013/02:47:16.8 to 12-May-2013/03:09:00.9 (UTC)
We will eventually concatenate the six datasets used here into one large dataset. However, we will keep them separate for now, as some of the steps to follow require individual datasets to be calibrated separately (namely, the sky/Tsys calibration and baseline subtraction). We therefore start by defining an array "basename" that includes the names of the six files in chronological order. This will simplify the following steps by allowing us to loop through the files using a simple for-loop in python. Remember that if you log out of CASA, you will have to re-issue this command. We will remind you of this in the relevant sections by repeating the command at the start.
# In CASA
basename=['uid___A002_X60b415_X39a','uid___A002_X60b415_X6f7','uid___A002_X6218fb_X264', 'uid___A002_X6218fb_X425','uid___A002_X6321c5_X3a7','uid___A002_X6321c5_X5ca']
Create Measurement Sets
The raw data have been provided to you in the ASDM format. ASDM stands for ALMA Science Data Model. It is the native format of the data produced by the observatory.
Before we can proceed to the calibration, we will need to convert those data to the CASA MS format. This is done simply with the task importasdm.
#In CASA
for name in basename:
importasdm(asdm = name)
Note: importasdm has an option singledish, which you may be tempted to use. It works, but it has some limitations (which will be removed in the future), so for now, we recommend not using it.
Initial Inspection
The usual first step is then to get some basic information about the data. We do this using the task listobs, which will output a detailed summary of each dataset supplied.
# In CASA
for name in basename:
listobs(vis=name+'.ms')
Note that after cutting and pasting a for-loop you often have to press return several times to execute. The output will be sent to the CASA logger. You will have to scroll up to see the individual output for each of the six datasets. Here is an example of the most relevant output for the fifth file in the list.
These commands define a python list called "basename", which contains the name of all 6 MS files. The "for" loop executes for each element in basename, calling listobs and directing the output to a file called, e.g., "uid___A002_X60b415_X39a.ms.listobs.txt" for the first measurement set. You can browse through the listobs output as you would normally look at a text file (use emacs, vi, or another editor). You can also send the output to the terminal from inside of CASA. To do so type:
# In CASA
os.system('cat uid___A002_X60b415_X39a.ms.listobs.txt')
or
# In CASA
os.system('more uid___A002_X60b415_X39a.ms.listobs.txt')
CASA knows a few basic shell commands like 'cat', 'ls', and 'rm' but for more complex commands you will need to run them inside 'os.system("command")'. For more information see http://casa.nrao.edu/ .
Here is an example of the (abridged) output from listobs for the first dataset in the list, uid___A002_X60b415_X39a.ms. You would see this if you had specified verbose to be False in the listobs call:
Observation: ALMA
Data records: 16588 Total integration time = 1302.91 seconds
Observed from 12-May-2013/02:22:16.9 to 12-May-2013/02:43:59.8 (UTC)
ObservationID = 0 ArrayID = 0
Date Timerange (UTC) Scan FldId FieldName nRows nUnflRows SpwIds Average Interval(s) ScanIntent
12-May-2013/02:22:16.3 - 02:22:42.8 1 0 M100 900 0.00 [0, 1, 2, 3, 4, 5, 6, 7, 8] [1.15, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHE
RE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE
02:23:24.3 - 02:25:11.4 2 0 M100 1524 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:25:25.2 - 02:27:11.2 3 0 M100 1522 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:27:25.1 - 02:29:11.0 4 0 M100 1522 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:29:24.9 - 02:31:12.0 5 0 M100 1524 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:31:25.8 - 02:33:11.8 6 0 M100 1522 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:34:03.6 - 02:34:29.0 7 0 M100 900 0.00 [0, 1, 2, 3, 4, 5, 6, 7, 8] [1.15, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48, 0.48] CALIBRATE_ATMOSPHE
RE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE
02:34:42.8 - 02:36:30.0 8 0 M100 1524 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:36:43.8 - 02:38:29.8 9 0 M100 1524 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:38:43.6 - 02:40:30.7 10 0 M100 1524 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:40:44.5 - 02:42:30.5 11 0 M100 1524 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
02:42:44.4 - 02:44:00.4 12 0 M100 1078 0.00 [0, 9, 10, 11, 12, 13, 14, 15, 16] [1.15, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01, 1.01] OBSERVE_TAR
GET#OFF_SOURCE,CALIBRATE_WVR#OFF_SOURCE
(nRows = Total number of rows per scan)
Fields: 1
ID Code Name RA Decl Epoch SrcId nRows nUnflRows
0 none M100 12:22:54.899040 +15.49.20.57200 J2000 0 16588 0.00
Spectral Windows: (17 unique spectral windows and 2 unique polarization setups)
SpwID Name #Chans Frame Ch1(MHz) ChanWid(kHz) TotBW(kHz) BBC Num Corrs
0 WVR#NOMINAL 4 TOPO 184550.000 1500000.000 7500000.0 0 XX
1 ALMA_RB_03#BB_1#SW-01#FULL_RES 128 TOPO 101942.187 -15625.000 2000000.0 1 XX YY
2 ALMA_RB_03#BB_1#SW-01#CH_AVG 1 TOPO 100926.562 1781250.000 1781250.0 1 XX YY
3 ALMA_RB_03#BB_2#SW-01#FULL_RES 128 TOPO 103757.337 -15625.000 2000000.0 2 XX YY
4 ALMA_RB_03#BB_2#SW-01#CH_AVG 1 TOPO 102741.712 1781250.000 1781250.0 2 XX YY
5 ALMA_RB_03#BB_3#SW-01#FULL_RES 128 TOPO 111814.962 15625.000 2000000.0 3 XX YY
6 ALMA_RB_03#BB_3#SW-01#CH_AVG 1 TOPO 112783.712 1781250.000 1781250.0 3 XX YY
7 ALMA_RB_03#BB_4#SW-01#FULL_RES 128 TOPO 113689.962 15625.000 2000000.0 4 XX YY
8 ALMA_RB_03#BB_4#SW-01#CH_AVG 1 TOPO 114658.712 1781250.000 1781250.0 4 XX YY
9 ALMA_RB_03#BB_1#SW-01#FULL_RES 4080 TOPO 101945.850 -488.281 1992187.5 1 XX YY
10 ALMA_RB_03#BB_1#SW-01#CH_AVG 1 TOPO 100949.756 1992187.500 1992187.5 1 XX YY
11 ALMA_RB_03#BB_2#SW-01#FULL_RES 4080 TOPO 103761.000 -488.281 1992187.5 2 XX YY
12 ALMA_RB_03#BB_2#SW-01#CH_AVG 1 TOPO 102764.906 1992187.500 1992187.5 2 XX YY
13 ALMA_RB_03#BB_3#SW-01#FULL_RES 4080 TOPO 111811.300 488.281 1992187.5 3 XX YY
14 ALMA_RB_03#BB_3#SW-01#CH_AVG 1 TOPO 112806.906 1992187.500 1992187.5 3 XX YY
15 ALMA_RB_03#BB_4#SW-01#FULL_RES 4080 TOPO 113686.300 488.281 1992187.5 4 XX YY
16 ALMA_RB_03#BB_4#SW-01#CH_AVG 1 TOPO 114681.906 1992187.500 1992187.5 4 XX YY
Sources: 19
ID Name SpwId RestFreq(MHz) SysVel(km/s)
0 M100 0 - -
0 M100 17 - -
0 M100 18 - -
0 M100 1 - -
0 M100 2 - -
0 M100 3 - -
0 M100 4 - -
0 M100 5 - -
0 M100 6 - -
0 M100 7 - -
0 M100 8 - -
0 M100 9 100950 0
0 M100 10 100950 0
0 M100 11 102794.1 0
0 M100 12 102794.1 0
0 M100 13 112794.1 0
0 M100 14 112794.1 0
0 M100 15 114669.1 0
0 M100 16 114669.1 0
Antennas: 2:
ID Name Station Diam. Long. Lat. Offset from array center (m) ITRF Geocentric coordinates (m)
East North Elevation x y z
0 PM01 T704 12.0 m -067.45.16.2 -22.53.22.1 42.8992 -520.1885 22.2159 2225113.044955 -5440122.820877 -2481517.728410
1 PM04 T703 12.0 m -067.45.16.2 -22.53.23.9 42.8809 -575.6904 21.7744 2225104.701420 -5440102.470120 -2481568.688227
This output shows that three sources were observed in each data set: M100.
- M100 are our science target. Note that the source corresponds to a number of individual fields (see the Field ID column).
The output also shows that the data contain many spectral windows. Using the labeling scheme in the listobs above these are:
- spw 9,spw 11,spw 13 and spw 15 hold our science data. These are "Frequency Domain Mode" (FDM) data with small (0.49 MHz) channel width and wide total bandwidth. As a result these have a lot of channels (4080). spw 15 holds the lower sideband (LSB) data and includes the CO(1-0) line.
We will focus on these data.
- spw 1,spw 3,spw 5 and spw 7 hold lower a resolution processing ("Time Domain Mode", TDM) of the data from the same part of the spectrum (baseband). These data have only 128 channels across 2 GHz bandwidth and so have a much coarser channel spacing than the FDM data.
These were used to generate the calibration tables that we include in the tarball but will not otherwise appear in this guide.
We do some initial inspection of the data using plotms. First we will plot amplitude versus channel, averaging over time in order to speed up the plotting process.
# In CASA
plotms(vis='uid___A002_X60b415_X39a.ms', xaxis='channel', yaxis='amp',
averagedata=T, avgtime='1e8', avgscan=T, iteraxis='antenna')
Next, we will look at amplitude versus time, averaging over channels and colorizing by field.
# In CASA
plotms(vis='uid___A002_X60b415_X39a.ms', xaxis='time', yaxis='amp',
averagedata=T, avgchannel='100', iteraxis='spw', coloraxis='field')
Convert MS format to single-dish format
In order to calibrate the data we need the data to be in the single-dish ASAP format. Most of the tasks that we will use for calibration are part of the ASAP package, which has been incorporated into CASA. The ASAP package uses a different data format, so from a global point of view, what we are going to do is, first convert the MS to the ASAP format, then run the necessary calibration tasks, then convert the data back to the MS format. Another important difference with interferometric data reduction is that the calibration is performed directly on the dataset, we will not produce calibration tables and apply them at the end. An effort is on-going to update the SD routines so that this is done, that should be available soon, but until then, please remember that all SD calibration operations apply to the data directly, so you may want to always create a new dataset each time, so that you do not have to start all over again.
We are now going to go through each of them with a bit more explanation. We will take the example of uid___A002_X6218fb_X264.ms.
We convert the data to the ASAP format using the routine sd.splitant for that. An outprefix has to be specified because the routine will create a dataset per antenna, taking the outprefix, and appending the antenna name and the format extension ('.asap').
# In CASA
sd.splitant(filename = 'uid___A002_X6218fb_X264.ms',
outprefix = 'uid___A002_X6218fb_X264.ms',
overwrite = True,
freq_tolsr = True)
The reason for the option freq_tolsr=True is to convert the frequencies in the MS from TOPO to LSRK. This is an important setting, as due to current limitations, it is the only stage where this conversion can be done. Using freq_tolsr=False would keep the frame of the frequencies from the MS, and would force you to image the final cube in TOPO velocities. Another important aspect is for the baselining/line finding: if your observations have been obtained at different epochs, it may be easier to work on spectra with LSRK frequencies because then the line emissions of all spectra can be overlapped.
We have now two ASAP datasets, one for PM03 and one for PM04. As usual, we will first obtain information about the content of the datasets, using sdlist (equivalent of listobs).
# In CASA
sdlist(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap',
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.sdlist')
sdlist(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap',
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.sdlist')
Here is an example of the most relevant output for uid___A002_X6218fb_X264.ms.PM04.asap.sdlist.
# In CASA
os.system('more uid___A002_X6218fb_X264.ms.PM04.asap.sdlist')
--------------------------------------------------------------------------------
Scan Table Summary
--------------------------------------------------------------------------------
Project: uid://A002/X5d9e5c/X42
Obs Date: 2013/04/28/04:10:19
Observer: cvlahakis
Antenna Name: ALMA//PM04@T703
Data Records: 15186 rows
Obs. Type: CALIBRATE_ATMOSPHERE#ON_SOURCE,CALIBRATE_WVR#ON_SOURCE
Beams: 1
IFs: 17
Polarisations: 2 (linear)
Channels: 4080
Flux Unit: K
Abscissa: Channel
Selection: none
Scan Source Time range Int[s] Record SrcType FreqIDs MolIDs
Beam Position (J2000)
--------------------------------------------------------------------------------
0 M100 2013/04/28/04:12:06.1 - 04:13:17.3 0.500364 594 [PSON:CALON, PSOFF:CALON] [0, 1, 2, 3, 4, 5, 6, 7, 8] [0]
0 J2000 12:23:07.0 +15.51.15.9
1 M100 2013/04/28/04:14:05.0 - 04:15:50.8 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.9 +15.51.17.4
2 M100 2013/04/28/04:16:09.4 - 04:17:56.4 1.01628 1443 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.8 +15.51.19.1
3 M100 2013/04/28/04:18:13.8 - 04:20:00.8 1.01628 1443 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.8 +15.51.20.6
4 M100 2013/04/28/04:20:19.4 - 04:22:05.2 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.7 +15.51.22.2
5 M100 2013/04/28/04:22:23.8 - 04:24:09.7 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.6 +15.51.23.7
6 M100 2013/04/28/04:25:03.7 - 04:26:14.7 0.500364 594 [PSON:CALON, PSOFF:CALON] [0, 1, 2, 3, 4, 5, 6, 7, 8] [0]
0 J2000 12:23:06.5 +15.51.25.6
7 M100 2013/04/28/04:26:33.8 - 04:28:19.6 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.5 +15.51.26.7
8 M100 2013/04/28/04:28:38.2 - 04:30:25.2 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.4 +15.51.28.1
9 M100 2013/04/28/04:30:42.6 - 04:32:29.6 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.3 +15.51.29.5
10 M100 2013/04/28/04:32:48.2 - 04:34:34.0 1.01619 1442 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.2 +15.51.30.9
11 M100 2013/04/28/04:34:52.6 - 04:36:08.5 1.0162 1018 [PSOFF, PSON] [0, 9, 10, 11, 12, 13, 14, 15, 16] [0, 1, 2, 3, 4]
0 J2000 12:23:06.2 +15.51.32.3
--------------------------------------------------------------------------------
FREQUENCIES: 9
ID IFNO Frame RefVal RefPix Increment Channels POLNOs
0 0 LSRK 1.87675e+11 1.5 2.5e+09 4 [0]
1 1 LSRK 1.0095e+11 63.5 -15625000 128 [0, 1]
2 2 LSRK 1.0092656e+11 0 -1.78125e+09 1 [0, 1]
3 3 LSRK 1.0276515e+11 63.5 -15625000 128 [0, 1]
4 4 LSRK 1.0274171e+11 0 -1.78125e+09 1 [0, 1]
5 5 LSRK 1.1280715e+11 63.5 15625000 128 [0, 1]
6 6 LSRK 1.1278371e+11 0 1.78125e+09 1 [0, 1]
7 7 LSRK 1.1468215e+11 63.5 15625000 128 [0, 1]
8 8 LSRK 1.1465871e+11 0 1.78125e+09 1 [0, 1]
9 9 LSRK 1.0095e+11 2039.5 -488281.25 4080 [0, 1]
10 10 LSRK 1.0094976e+11 0 -1.9921875e+09 1 [0, 1]
11 11 LSRK 1.0276515e+11 2039.5 -488281.25 4080 [0, 1]
12 12 LSRK 1.0276491e+11 0 -1.9921875e+09 1 [0, 1]
13 13 LSRK 1.1280715e+11 2039.5 488281.25 4080 [0, 1]
14 14 LSRK 1.1280691e+11 0 1.9921875e+09 1 [0, 1]
15 15 LSRK 1.1468215e+11 2039.5 488281.25 4080 [0, 1]
16 16 LSRK 1.1468191e+11 0 1.9921875e+09 1 [0, 1]
--------------------------------------------------------------------------------
MOLECULES:
ID RestFreq Name
0 [] []
1 [1.0095e+11] [Manual_window(ID=0)]
2 [1.02794e+11] [Manual_window(ID=0)]
3 [1.12794e+11] [Manual_window(ID=0)]
4 [1.14669e+11] [CO_v_0_1_0(ID=3768098)]
--------------------------------------------------------------------------------
Calibration
About the calibration itself: the two main steps are
1. Calibration of the spectra into K, by applying the Tsys calibration and removing the signal from the OFF position 2. Baselines Subtraction (i.e. subtracting the background emission, to keep only the line emission.)
Tsys Correction
Let's start by checking the Tsys solutions. We will use the gencal command used for interferometric data reduction. This will produce a CASA calibration table, which can be analysed similarly to previous guides M100 Band3 ACA 4.1#Tsys
# In CASA
gencal(vis = 'uid___A002_X6218fb_X264.ms',
caltable = 'uid___A002_X6218fb_X264.ms.tsys',
caltype = 'tsys')
plotbandpass(caltable='uid___A002_X6218fb_X264.ms.tsys', overlay='time',
xaxis='freq', yaxis='amp', subplot=22, buildpdf=False, interactive=False,
showatm=True,pwv='auto',chanrange='5~123',showfdm=True,
field='', figfile='uid___A002_X6218fb_X264.ms.tsys.plots.overlayTime/uid___A002_X6218fb_X264.ms.tsys')
This sequence loops over all of our files and plots Tsys as a function of time for channel. In the call to plotcal:
- subplot=22 parameter sets up a 2 x 2 panel grid.
- iteration tells plotcal to make a separate plot for each antenna.
<figure id="X264.Tsys.png">
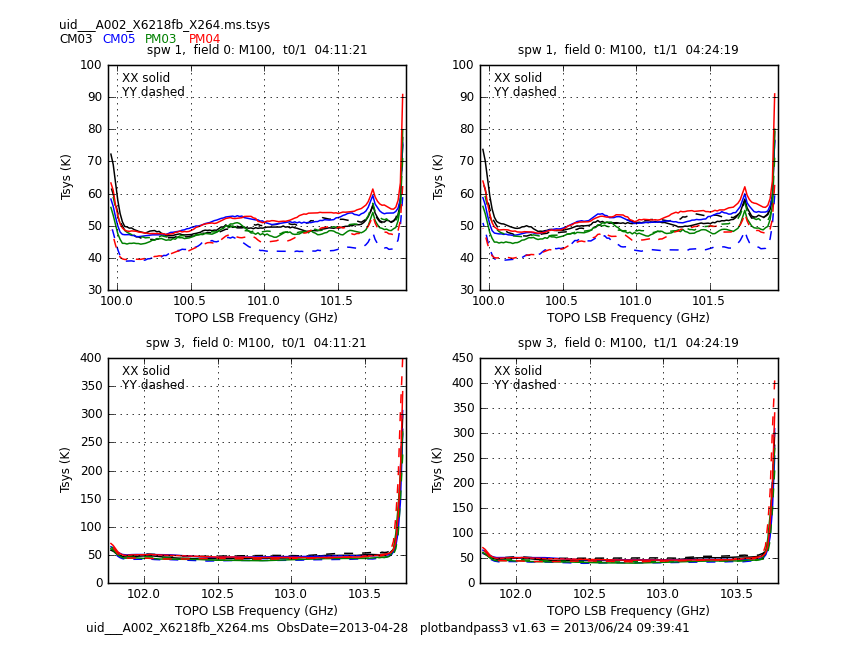
</figure>
Fill Tsys Column
For the spectral window association, we will use the routine tsysspwmap available directly in CASA.
# In CASA
from recipes.almahelpers import tsysspwmap
tsysmap = tsysspwmap(vis = 'uid___A002_X6218fb_X264.ms', tsystable = 'uid___A002_X6218fb_X264.ms.tsys')
tsysmap is a list of indices, with tsysmap[i] being the index of the Tsys spw to associate to the science spw of index i.
In the next step, we will do the actual preparation of the Tsys (i.e. re-sampling and re-indexing of the Tsys spw), using the library filltsys, also available directly in CASA.
# In CASA
import filltsys
for i in [9, 11, 13, 15]:
filltsys.fillTsys('uid___A002_X6218fb_X264.ms.PM03.asap',
specif = i,
tsysif = tsysmap[i],
mode = 'linear',
extrap = True)
for i in [9, 11, 13, 15]:
filltsys.fillTsys('uid___A002_X6218fb_X264.ms.PM04.asap',
specif = i,
tsysif = tsysmap[i],
mode = 'linear',
extrap = True)
This will loop through each science spw, re-sample the Tsys solutions associated to spw of index tsysmap[i] using a linear interpolation, allowing for extrapolation in case the science and Tsys spw do not perfectly match, and re-index it to spw of index i. You will note that the SD tasks use the word 'if': this is equivalent to 'spw id' in interferometry tasks.
View Spectra
# In CASA
sdplot(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap',
iflist = [15],
scanaverage = True,
plottype = 'spectra',
stack = 'pol',
panel = 'scan',
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.png',
overwrite = True)
A Priori Flagging
We have now prepared the Tsys solutions for application. We have not applied them yet. We will first do some a-priori flagging, of the edge channels. This is similar to flagging edge channels in TDM interferometry datasets. In FDM interferometry datasets, these edge channels are not written out, so there is no need to flag anything. Here, we have an FDM SD dataset, produced with the second correlator (the ACA correlator), which does not excise them (yet).
Each spw covers the full baseband width (2GHz), so we will flag 120 channels on each side so as to keep only the center 3080 channels, similarly to FDM interferometry datasets.
# In CASA
sdflag(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap',
specunit = 'channel',
iflist = [9, 11, 13, 15],
maskflag = [[0, 119], [3960, 4079]],
overwrite = True)
sdflag(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap',
specunit = 'channel',
iflist = [9, 11, 13, 15],
maskflag = [[0, 119], [3960, 4079]],
overwrite = True)
If you want to flag data in tool base, we will present an example for that.
# In CASA
param_org = sd.rcParams['scantable.storage']
sd.rcParams['scantable.storage'] = 'disk'
s = sd.scantable('uid___A002_X6218fb_X264.ms.PM03.asap', average=False)
sel = sd.selector()
sel.set_ifs([9, 11, 13, 15])
s.set_selection(sel)
mask = s.create_mask([[0, 119], [3960, 4079]])
s.flag(mask)
s.set_selection()
del s
sd.rcParams['scantable.storage'] = param_org
Apply Calibration and Inspect
Now is the step where we will actually calibrate the data. This is done with the task sdcal. This is a straight-forward operation, so no opportunity for tweaking. The calibration mode (calmode) shall be 'ps', as in Position Switching. The task will find automatically the OFF-position measurements. We need to specify the science spws (iflist) because they are the only ones that can be calibrated. We specify a different output file (outfile) so as to be able to come back to the original dataset if necessary.
# In CASA
sdcal(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap',
calmode = 'ps',
iflist = [9, 11, 13, 15],
scanaverage = False,
timeaverage = False,
polaverage = False,
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal',
overwrite = True)
sdcal(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap',
calmode = 'ps',
iflist = [9, 11, 13, 15],
scanaverage = False,
timeaverage = False,
polaverage = False,
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal',
overwrite = True)
Before proceeding to the next step, we can plot the calibrated spectra, using the sdplot task. The commands below will plot one spectrum per scan, spw and polarization. It is difficult to describe what a good spectrum looks like at this stage. What you should look for is outliers (i.e. spectra that differ from most, whether in shape or level).
# In CASA
for i in [9, 11, 13, 15]:
sdplot(infile='uid___A002_X6218fb_X264.ms.PM03.asap.cal',
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan',
outfile='uid___A002_X6218fb_X264.ms.PM03.asap.cal.plots/uid___A002_X6218fb_X264.ms.PM03.asap.cal.spectra.spw'+str(i)+'.png',
overwrite = True)
for i in [9, 11, 13, 15]:
sdplot(infile='uid___A002_X6218fb_X264.ms.PM04.asap.cal',
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan',
outfile='uid___A002_X6218fb_X264.ms.PM04.asap.cal.plots/uid___A002_X6218fb_X264.ms.PM04.asap.cal.spectra.spw'+str(i)+'.png',
overwrite = True)
View Calibrated Spectra
# In CASA
sdplot(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal',
iflist = [15],
scanaverage = True,
plottype = 'spectra',
stack = 'pol',
panel = 'scan',
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.png',
overwrite = True)
<figure id="X264.PM04.cal.png">
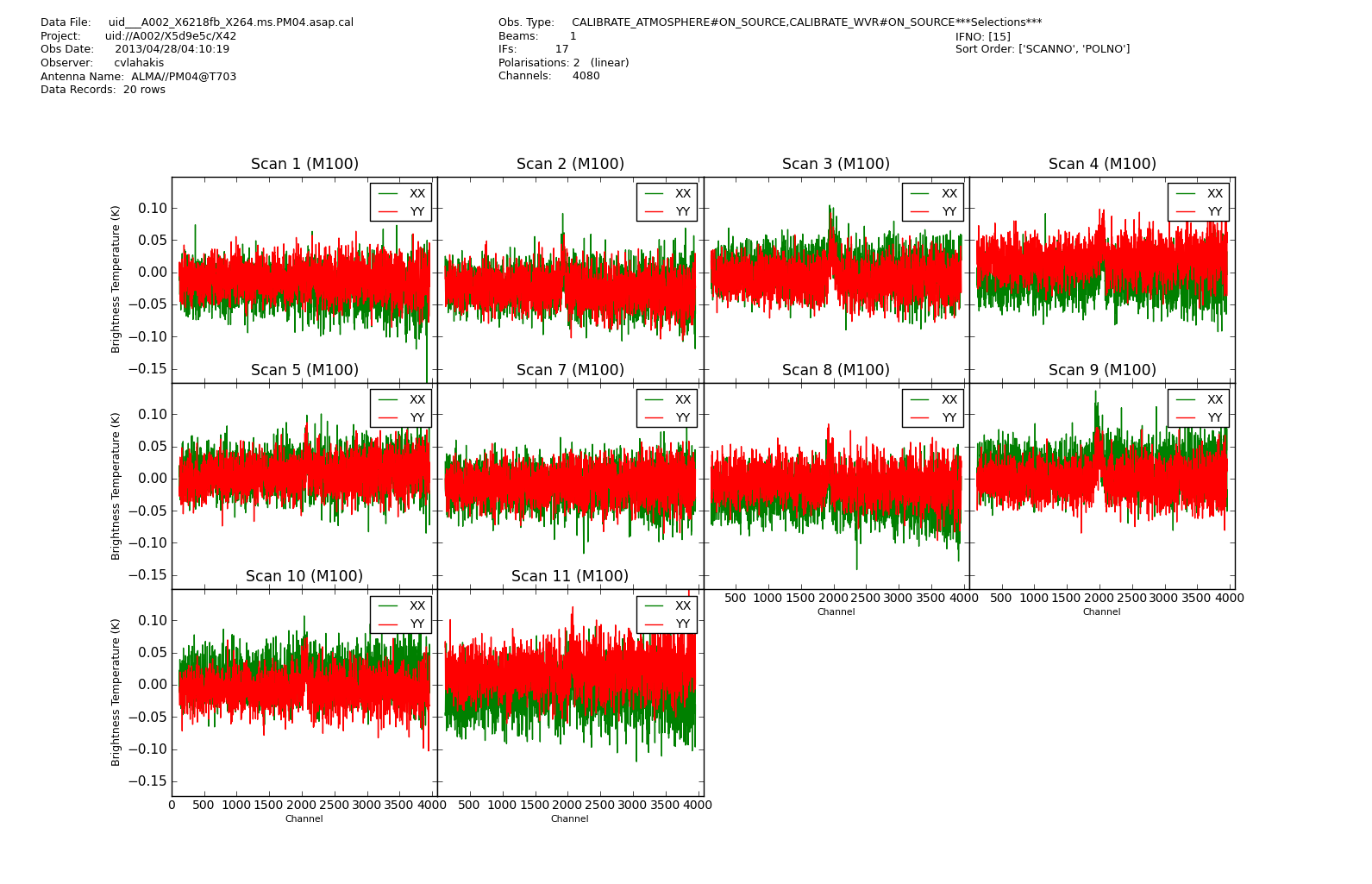
</figure>
Baseline Subtraction and Inspect
We will now perform the second main step of the calibration: the baselining. This is done with the sdbaseline task. In the commands below, the baseline fitting method is controlled by the parameters blfunc and order. Many fitting methods are available (’poly’,’chebyshev’,’cspline’,’sinusoid’, see help of sdbaseline task for more information). Here, we will do a simple linear fitting.
The parameter maskmode allows excluding portions of the spectra from the baseline fitting. This is mainly useful to exclude channels where there are line emissions (it could also be used to excluse the edge channels, in case they were not flagged beforehand). The channels can be specified as a list or chosen interactively. Another option is to use the line finder implemented in the sdbaseline task, using maskmode = 'auto'; then the parameters thresh and avg_limit can be used to tweak the line finding algorithm. (thresh is specified in sigma units, avg_limit is specified as a number of channels.)
# In CASA
sdbaseline(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal',
iflist = [9, 11, 13, 15],
maskmode = 'auto',
thresh = 3.0,
avg_limit = 8,
blfunc = 'poly',
order = 1,
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl',
overwrite = True)
sdbaseline(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal',
iflist = [9, 11, 13, 15],
maskmode = 'auto',
thresh = 3.0,
avg_limit = 8,
blfunc = 'poly',
order = 1,
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl',
overwrite = True)
We run again sdplot to check the output spectra, fully calibrated this time.
# In CASA
for i in [9, 11, 13, 15]:
sdplot(infile='uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl',
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan',
outfile='uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.plots/uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.spectra.spw'+str(i)+'.png',
overwrite = True)
for i in [9, 11, 13, 15]:
sdplot(infile='uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl',
iflist=[i], plottype='spectra', specunit='channel', scanaverage=True, stack='pol', panel='scan',
outfile='uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl.plots/uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl.spectra.spw'+str(i)+'.png',
overwrite = True)
We have now entirely calibrated the dataset Into Kelvin units. The conversion to Jy will be covered into the guide on combination.
View Baselined Spectra
# In CASA
sdplot(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl',
iflist = [15],
specunit = 'GHz',
scanaverage = True,
stack = 'pol',
panel = 'scan',
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.png',
overwrite = True)
<figure id="X264.PM04.cal.bl.png">
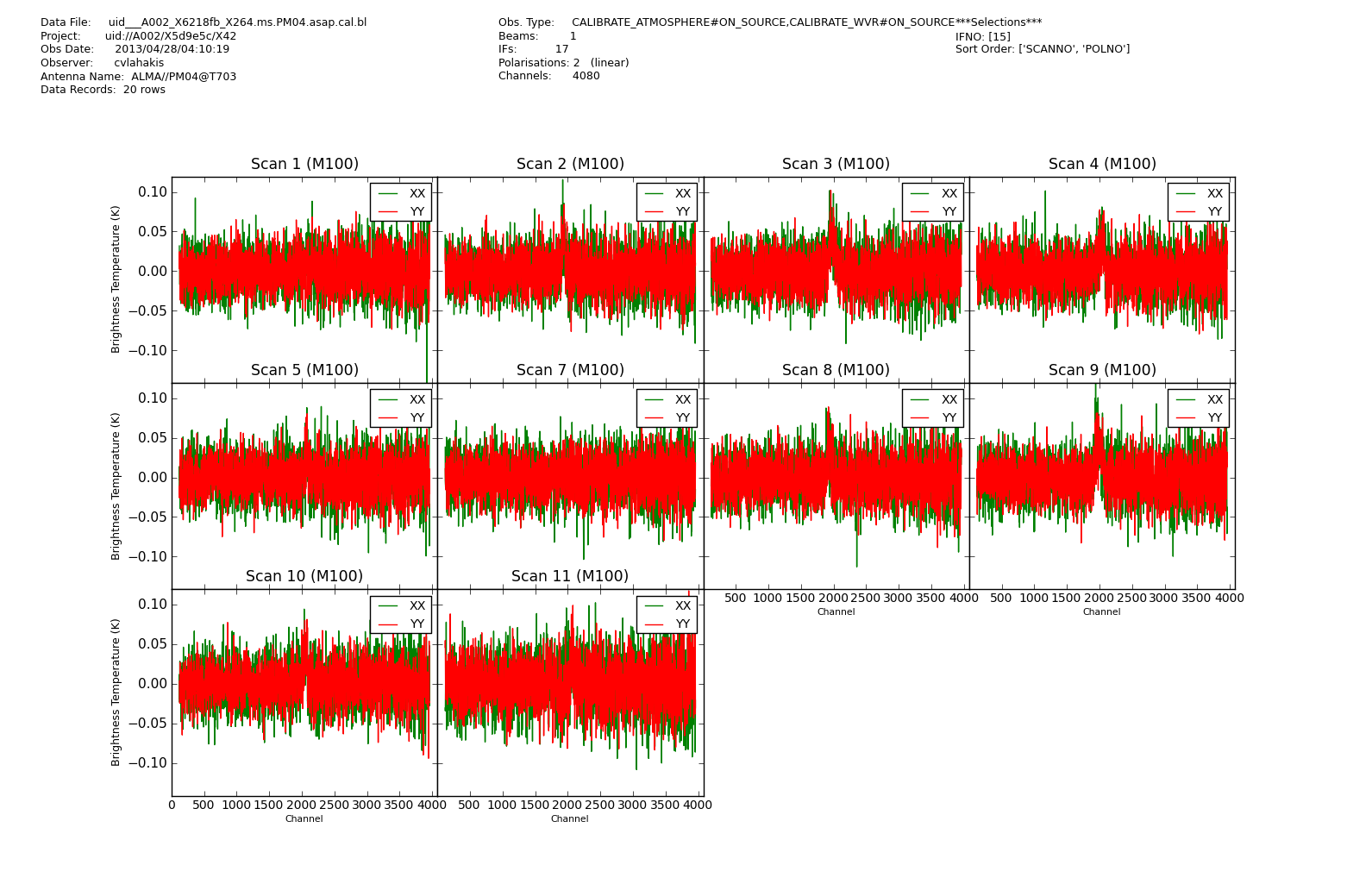
</figure>
Prepare for Imaging
Convert each ASAP dataset to an MS
Here, we will convert all of the calibrated asap dataset into measurement sets. The CASA task sdsave will do this.
#In CASA
sdsave(infile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl',
outfile = 'uid___A002_X6218fb_X264.ms.PM03.asap.cal.bl.ms',
outform = 'MS2')
sdsave(infile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl',
outfile = 'uid___A002_X6218fb_X264.ms.PM04.asap.cal.bl.ms',
outform = 'MS2')
Combine all executions to one MS
Concatenate all of the calibrated measurement sets into one for imaging. The CASA task concat will do this.
#In CASA
os.system('rm -rf concat_m100.ms')
concat(vis='uid___A002_X60b415_X39a.ms.cal.split', 'uid___A002_X60b415_X6f7.ms.cal.split', 'uid___A002_X6218fb_X264.ms.cal.split', 'uid___A002_X6218fb_X425.ms.cal.split', 'uid___A002_X6321c5_X3a7.ms.cal.split', 'uid___A002_X6321c5_X5ca.ms.cal.split'],
concatvis='concat_m100.ms',
freqtol='10MHz')
The individual calibrated MSs have slightly different observing frequencies, although the rest frequencies are the same. The freqtol parameter sets the tolerance for considering whether the different spectral windows from the input datasets should be output as the same spectral window ID.
Image the Total Power Data
Run listobs on the total power data to see what spw contains the CO(1-0).
#In CASA
os.system('rm -rf concat_m100.ms.listobs')
listobs(vis='concat_m100.ms',listfile='concat_m100.ms.listobs')
Spectral window SPWID=3 contains the 115.27 GHz line, so we image this window. The task sdimaging will do this.
#In CASA
os.system('rm -rf M100_SD_cube.image')
sdimaging(infile='concat_m100.ms',
field=0,spw=15,
specunit='km/s',restfreq='115.271204GHz',
dochannelmap=True,
nchan=70,start=1400,step=5,
gridfunction='gjinc',imsize=[50,50],
cell=['10arcsec','10arcsec'],
phasecenter = 'J2000 12h22m54.9 +15d49m15'
outfile='M100_SD_cube.image')
The restfreq parameter must be specified when using "km/s" as the units, as in this case. Start and step parameters are specified in units that the user chooses for specunit. The numbers here are chosen so that the resulting image has the same number of channels, velocity range and channel width as the 7m and 12m array images.
<figure id="X264.M100.chan.png">
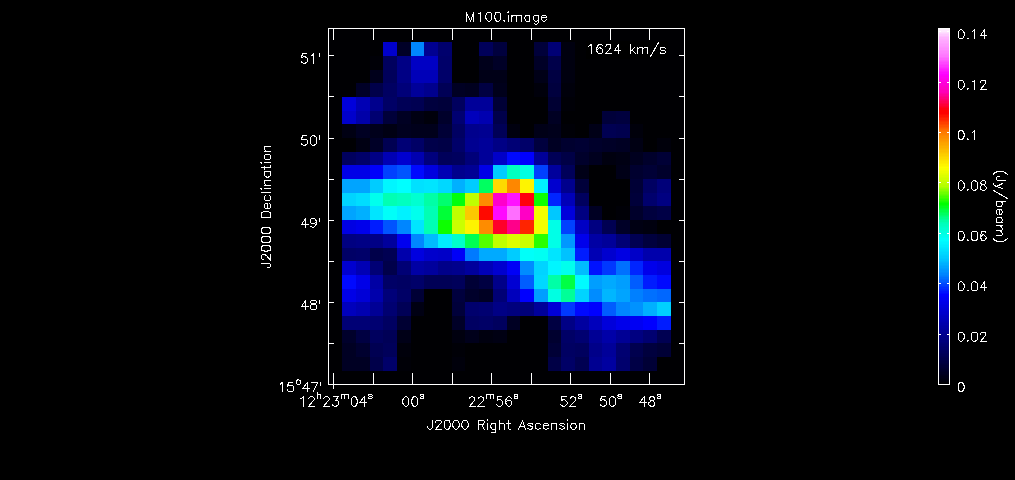
</figure>
Gridding Function
The gridfunction is the weighting function that is used to grid the observed flux to individual pixels in the image.
"SF": a spheroidal function, which minimizes aliasing effects. "BOX": a pillbox function, which defaults to a kernel box size of 1 pixel. "PB" (primary beam): assumes an Airy disk, corresponding to an antenna with 10.7m diameter, the effective diameter of an ALMA 12m antenna. "GAUSS": a gaussian, and its size can be defined by additional subparameters (truncate and gwidth). "GJINC": a gaussian convolved with the Bessel function, and can minimize the broadening of the effective beam.
Any of the functions which require the observing frequency for determining the beam size will read the frequency from the dataset, and the user can use the default. The cell size should be chosen so that it is about 1/3 to 1/4 of the FWHM of the effective beam.
Image Analysis : Moment Maps
Next we will make moment maps for the CO(1-0) emission: Moment 0 is the integrated intensity; Moment 1 is the intensity weighted velocity field; and Moment 2 is the intensity weighted velocity dispersion.
Above we determined the rms noise levels for M100 mosaics in a line-free and a line-bright channel. We want to limit the channel range of the moment calculations to those channels with significant emission. One good way to do this is to open the cube in the viewer overlaid with 3-sigma contours, with sigma corresponding to the line-free rms.
For moment 0 (integrated intensity) maps you do not typically want to set a flux threshold because this will tend to noise bias your integrated intensity.
# In CASA
os.system('rm -rf M100_SD_cube.image.mom*')
immoments(imagename = 'M100_SD_cube.image',
moments = [0],
axis = 'spectral',
chans = '1~24',
outfile = 'M100_SD_cube.image.mom0')
For higher order moments it is very important to set a conservative flux threshold. Typically something like 6sigma, using sigma from a bright line channel works well. We do this with the mask parameter in the commands below. When making multiple moments, immoments appends the appropriate file name suffix to the value of outfile.
# In CASA
os.system('rm -rf M100_SD_cube.image.mom*')
immoments(imagename = 'M100_SD_cube.image',
moments = [1],
axis = 'spectral',
chans = '1~24',
includepix = [0.018, 1.0],
outfile = 'M100_SD_cube.image.mom1')
Export data as fits
If you want to analyze the data using another software package it is easy to convert from CASA format to FITS.
# In CASA
os.system('rm -rf M100_SD_*.fits')
exportfits(imagename='M100_SD_cube.image', fitsimage='M100_SD_cube.image.fits')
exportfits(imagename='M100_SD_cube.image.mom0', fitsimage='M100_SD_mom0.fits')
Although "FITS format" is supposed to be a standard, in fact most packages expect slightly different things from a FITS image. If you are having difficulty, try setting velocity=T and/or dropstokes=T.
Continue on to Combining Images with 7m and 12m dataset
Now you can continue on to the M100_Band3_Combine_4.1.
Last checked on CASA Version 4.1.0.